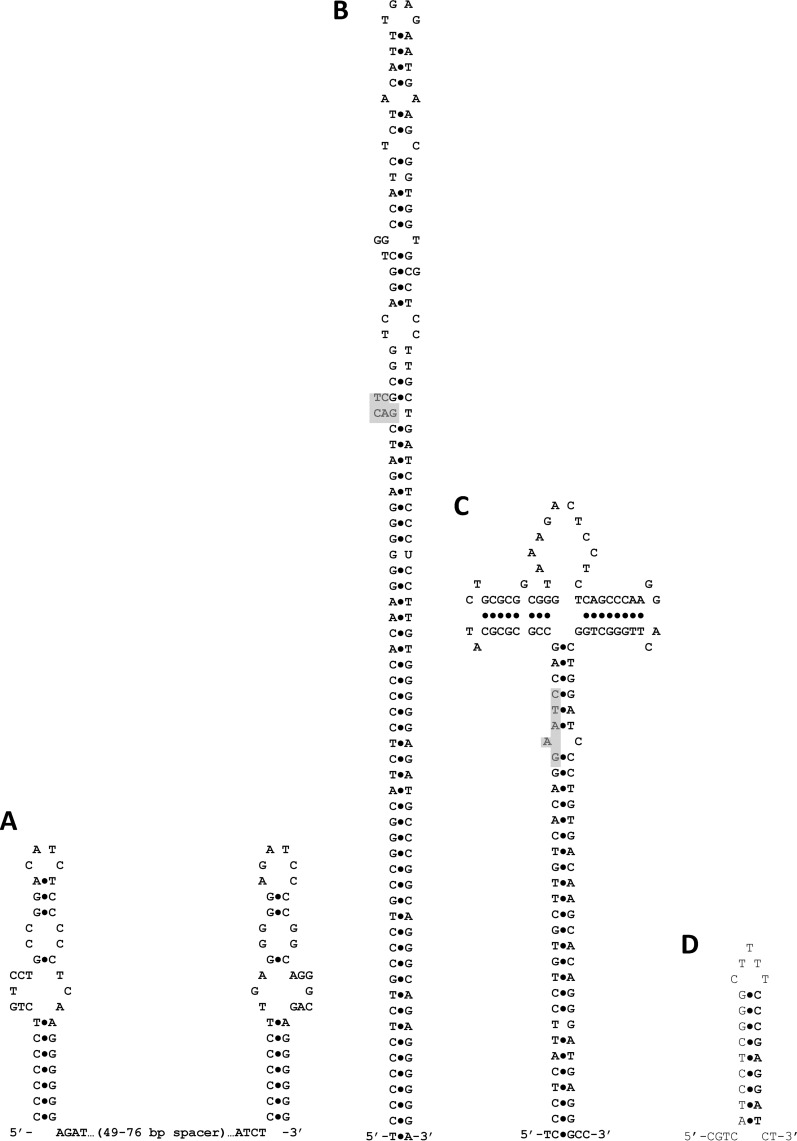
Fig 1.
Potential secondary structures for genomic repeat elements. (A) The structure emphasizes the inverted repeats present at each end of the CIR2 full element. (B) An alternative secondary structure for a CIR2 full element that emphasizes the larger overall inverted repeat. The extent of the stem forming above position 44 depends on the sequence of the spacer region. The shaded polygon indicates where a CcrM binding site sits if one is present (it is found in only about one-third of the elements). (C) Potential secondary structure for CIR1 elements. The structure emphasizes the larger overall inverted repeat. The shaded area indicates the usual location of a CcrM binding site, but quite often, a second CcrM binding site exists directly opposite the first location in a full element (the example here does not contain such a second site). (D) Potential secondary structure for KE3 half elements and ends of KE3 full elements.