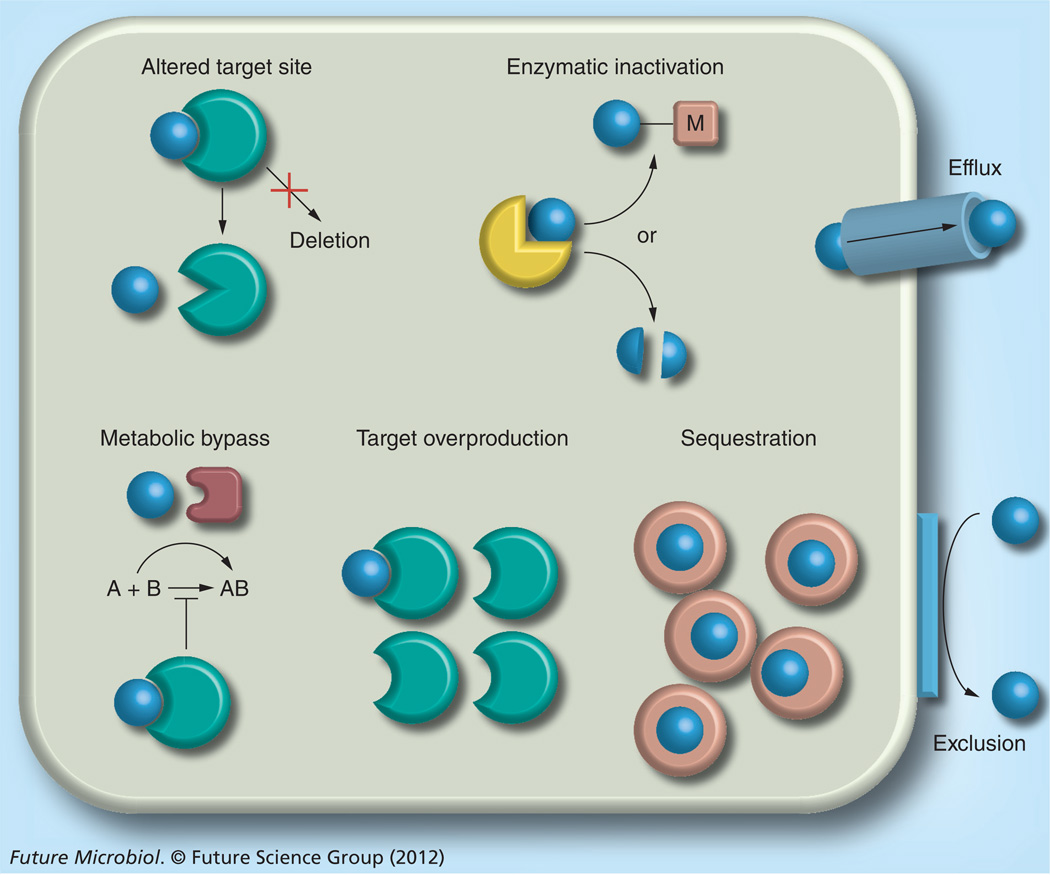
Figure 1. Bacterial antibiotic resistance mechanisms.
Bacterial antimicrobial resistance mechanisms are multifaceted, ranging from: exclusion of drug molecules (blue spheres) from the bacterial cell by physicochemical constraints (e.g., porins or lipopolysaccharide); efflux from the cell via active transport mechanisms; enzymatic inactivation, either in the form of substrate cleavage or chemical modification (M = acetylation, adenylation or phosphorylation); target site alteration or, rarely, target deletion; metabolic bypass by substitution of a susceptible enzyme or pathway with a resistant enzyme or pathway; target overproduction by either increased transcription or gene multiplication; and drug sequestration by specific binding proteins akin to immunity proteins. Details about individual resistance mechanisms and specific examples are provided in the text. Bacteria often employ different resistance mechanisms that act synergistically, for instance, efflux and exclusion, to achieve high-level resistance [26].