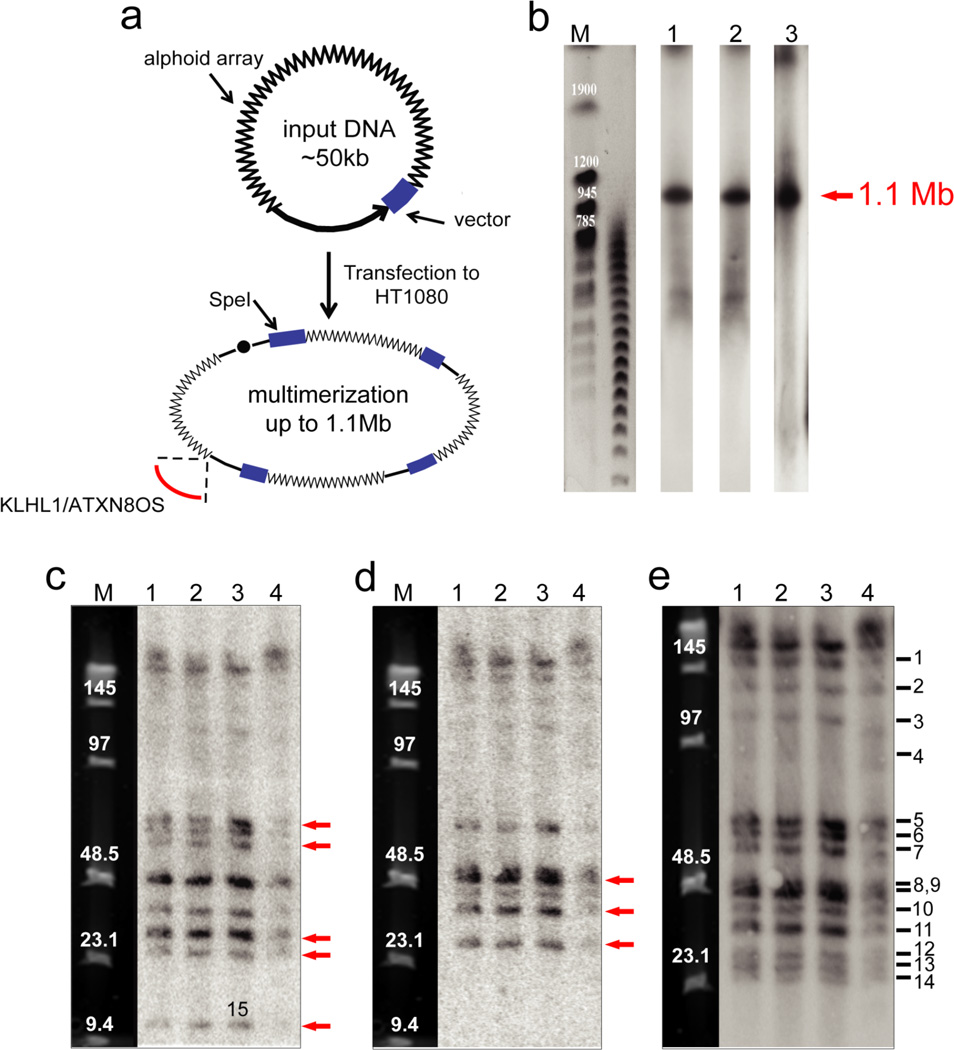
Figure 1.
Structural analysis by restriction enzyme digestion and CHEF of the alphoidtetO-HAC in human, hamster and chicken cells. (a) Diagram illustrating multimerization of input DNA during de novo HAC formation in human cells. Input DNA consists of 40 kb alphoid DNA and 10 kb vector sequence. Chromosome 13 fragment carrying the KLHL1/ATXN8OS genes was captured during formation of the alphoidtetO-HAC. (b) Determination of the size of the multimerized input DNA in the alphoidtetO-HAC in chicken DT40, hamster CHO and human HT1080 cells. Genomic DNA possessing the HAC was digested with PmeI and separated by CHEF gel electrophoresis (range 200–1500 kb). The transferred membranes were hybridized with a radioactively labeled tetO-specific alphoid probe. A single1.1 Mb fragment was detected. Its size was determined by comparison with the DNA size standard, S. cerevisiae chromosomes. Lane 1 – the HAC in chicken cells; lane 2 – the HAC in hamster cells; lane 3 – the HAC in human cells. (c) Analysis of the alphoidtetO-HAC digested by SpeI. Genomic DNA possessing the HAC was digested with SpeI endonuclease and separated by CHEF gel electrophoresis (range 10–70 kb). The transferred membranes were hybridized with either vector probe 3 (c), vector probe 1 (d) or tetO-alphoid (e) probe. Red arrows indicate to the fragments that are specific to either probe 1 or probe 3. Lane 1 – the HAC in DT40 cells; lane 2 - the HAC in CHO cells; lane 3 - the original alphoidtetO-HAC in human HT1080 cells; lane 4 - the HAC transferred back to human cells from CHO cells. M - Pulse Markers.