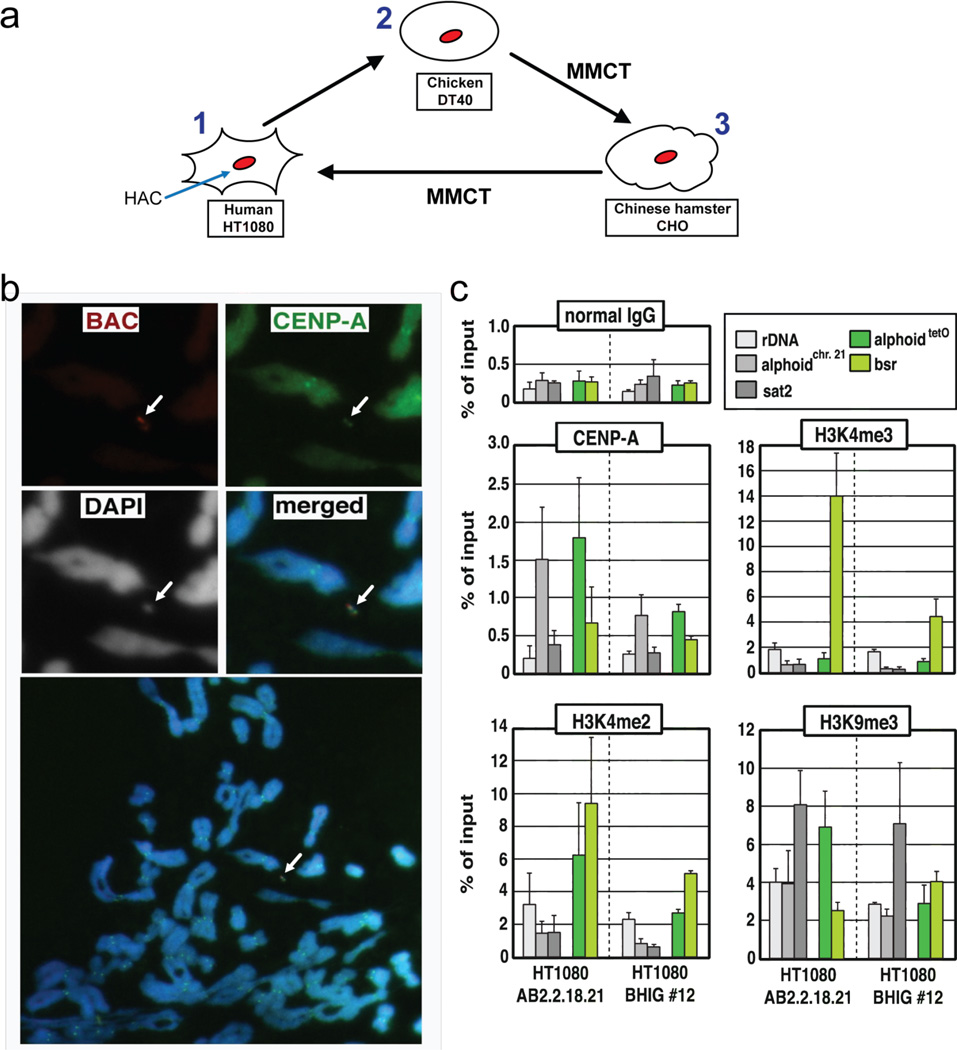
Figure 4.
Integrity of the functional domains in the alphoidtetO-HAC that underwent three rounds of MMCT transfer. (a) The alphoidtetO-HAC was first transferred to chicken DT40 from human HT1080. A loxP cassette was inserted into the HAC by homologous recombination in DT40 cells. Then the HAC was transferred to CHO cells. From CHO cells the HAC was transferred back to HT1080. (b) Immuno-FISH analysis of metaphase chromosome spreads containing the alphoidtetO-HAC in human HT1080 cells. Cells with the alphoidtetO-HAC (clone BHIG#12) were used for analysis. Immunolocalization of the centromeric protein CENP-A on metaphases was performed by indirect immunofluorescence with anti–CENP-A antibody and Alexa 488-conjugated secondary antibody (green). HAC-specific DNA sequence (RCA/SAT43 vector) was used as a FISH probe to detect the HAC (red). CENP-A and BAC signals on the HAC overlap one another. (c) ChIP analysis of CENP-A assembly and modified histone H3 at the alphoidtetO-HAC. The results of ChIP analysis using normal mouse IgG (top left panel), antibodies against CENP-A (middle left panel), dimethylated histone H3 Lys4 (H3K4me2, bottom left panel), trimethylated histone H3 Lys4 (H3K4me3, middle right panel) and trimethylated histone H3 Lys9 (H3K9me3, bottom right panel). The assemblies of these proteins on the original alphoidtetO-HAC in the AB2.2.18.21 cell line (left), the alphoidtetO-HAC in the BHIG#12 cell line (right) are shown. The bars show the percentage recovery of the various target DNA loci by immunoprecipitation with each antibody to input DNA. Error bars indicate s.d. (n= 2 or 3). Analyzed loci were rDNA (5S ribosomal DNA), alphoidchr.21 (centromeric alphoid DNA of chromosome 21), sat2 (pericentromeric satellite 2), alphoidtetO (alphoid DNA with tetO motif on tetO alphoid HAC), Bsr (the marker gene in BAC vector region of tetO alphoid HAC). Comparison of the enrichment of tetO-alphoid DNA and the endogenous chromosome 21 alphoid DNA by CENP-A, H3K4me2, H3K4me3 in BHIG#12 and AB2.2.18.21 cells was carried out by calculations of the ratio between IPed tetO-alphoid DNA in the HAC and IPed endogenous chromosome 21 alphoid DNA for each cell line. As seen in Figure S1 (Supporting Information), a relative enrichment of CENP-A, H3K4me2, H3K4me3 and H3K9me3 on tetO-alphoid DNAs in BHIG#12 cells is not different from that observed in AB2.2.18.21 cells. This indicates that kinetochore regions in the HAC did not change after multiple steps of HAC transfer via MMCT.