Fig. 2. PEGylation of AAP and toeprinting of AAP in tape-measure context.
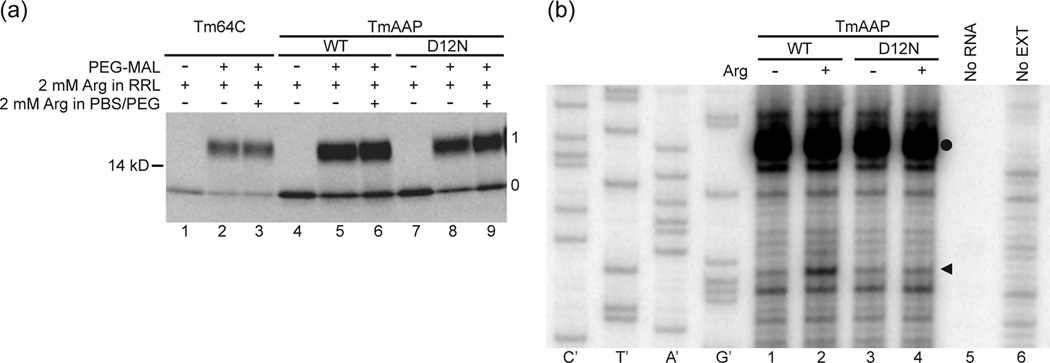
(a) Nascent peptides were PEGylated and fractionated on polyacrylamide gels as described in the Methods. A control construct, an all-extended tape measure with a reporter position at residue 64C (Tm, lanes 1–3) was compared to AAP substituted into the tape measure two residues from the PTC (TmAAP, lanes 4–6) and D12N AAP in the same location (TmAAP D12N, lanes 7–9). Lanes 1, 4, and 7 are derived from a sample not treated with PEG-MAL. All other lanes derive from samples incubated with 1mM PEG-MAL for 3h. Gels were 4–12% NuPAGE Bis-Tris gels with MOPS running buffer. The number to the left of the gel is a molecular weight standard; numbers to the right of the gel indicate unPEGylated (0) and singly PEGylated (1) protein. (b). WT TmAAP and D12N TmAAP mRNA were used to program reticulocyte lysates as indicated. Lysates were supplemented with 10 µM (−) or 3 mM (+) Arg and 10 µM each of the other 19 amino acids After 20 min of translation, 3 µl of translation mixtures were toeprinted with primer Tm. Lanes indicated as no RNA or no EXT show toeprinting of lysate without added RNA and RNA in the absence of lysate, respectively. The Tm primer was also used for dideoxynucleotide sequencing of WT TmAAP DNA template (leftmost sequencing markers). The arrowhead indicates the position of the toeprint corresponding to ribosomes with AAP codon-25 (a CCA Pro-codon in this construct) in the A site. The toeprint signal marked with a closed circle is also observed in translation reactions programmed with these mRNAs to which cycloheximide was added at time 0 and thus does not represent ribosomes stalled during translation of this region of TmAAP.
