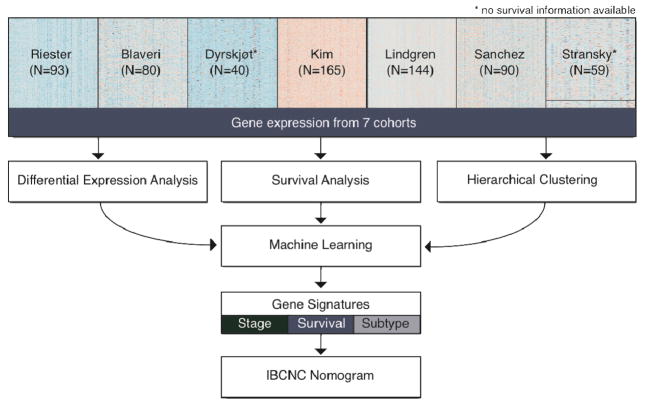
Figure 1. Overview of the study.
This study represents a massive meta-analysis of our cohort of high-risk bladder cancer patients (Table 1) and 6 independent studies (Table 2). All data was subject to comprehensive differential expression and survival analyses and to hierarchical clustering. Machine learning algorithms were used to find gene signatures predictive of stage, survival and molecular subtype. The survival gene signature was then included in a validated postoperative nomogram, developed by the International Bladder Cancer Nomogram Consortium (IBCNC) (28).