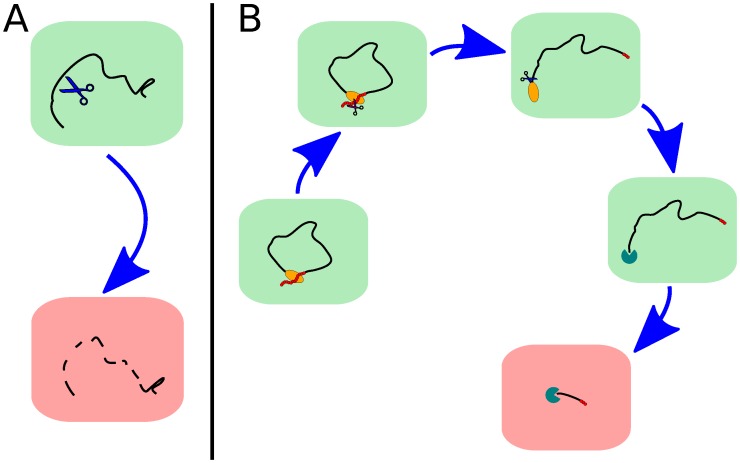
Figure 2. Prototypical pathways of mRNA degradation.
In panel A degradation is depicted as a relatively simple process determined by only a single step, e.g. by unspecific and fast endonucleolytic decay, such as the degradation pathway mediated by RNase E in prokaryotic cells. In panel B, instead, we show a schematic representation of the degradation pathway known as decapping which is one of the main degradation mechanisms in eukaryotic cells. The decapping mechanism consists of several biochemical steps, possibly triggered by a specific miRNA, which contribute to destabilize the mRNA until complete degradation takes place. This mechanism can be considered as a prototype of multi-step degradation.