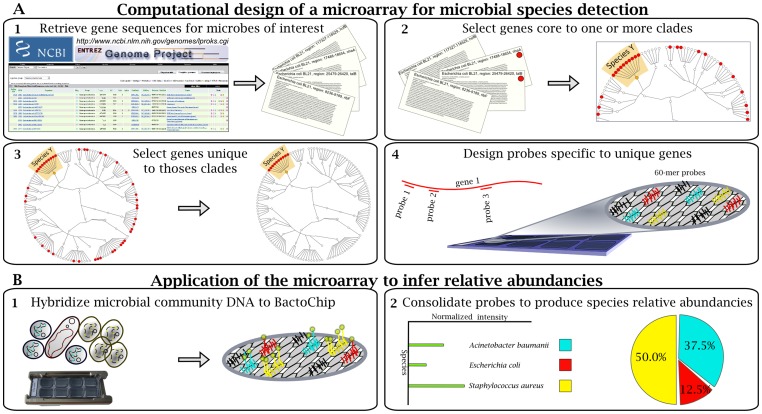
Figure 1. Design of the BactoChip and its application for microbial species identification and quantification.
(A) Schematic overview of computational design and its output. (A1) Complete genomes from 186 bacterial species were retrieved from the National Centre for Biotechnology Information microbial database. (A2) Gene sequences core to each target species were defined on the basis of sequence conservation within each clade. Red dots represent the distribution of core genes shared by strains within and outside a target clade. (A3) Core genes unique to each target species were selected by sequence alignment against all available archaeal and bacterial sequences. (A4) Oligonucleotide probes were designed for up to 10 identified unique genes for each target bacterial species. Each probe color represents specificity to a defined bacterial species. (B) Experimental design and example data. (B1) DNA from microbial communities was tested on the BactoChip. Green dots represent Cy3 bound to genomic DNA fragments from a sample hybridized to the chip. (B2) Species relative abundances are finally inferred by normalization of the fluorescence signal for each probe and species.