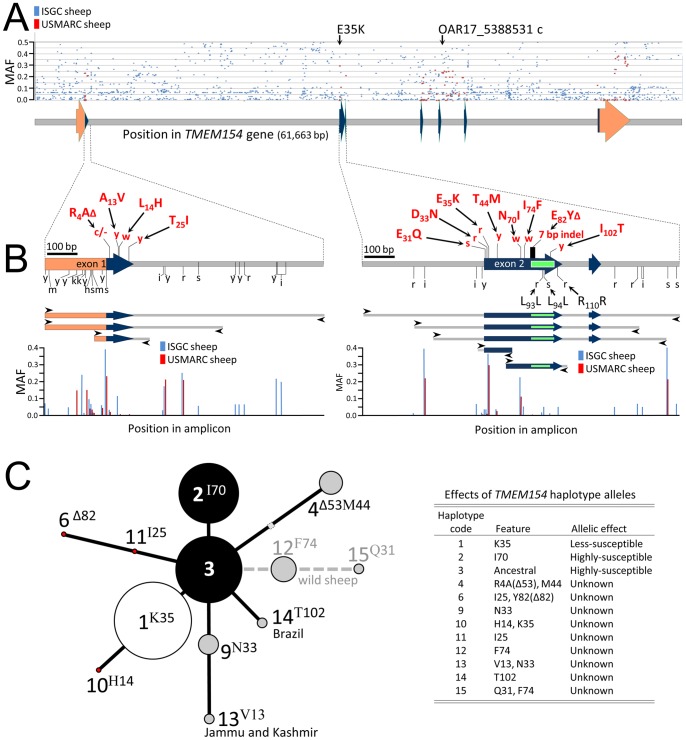
Figure 2. TMEM154 SNP maps and median-joining networks.
Panel A, genomic map of TMEM154: orange arrows, 5′ and 3′untranslated regions of exons; blue arrows, exon coding regions; grey rectangles, introns or intergenic regions. Blue and red tick dots denote position and frequency of SNPs in an international panel of 75 sheep and a panel of 96 U.S. sheep [9], respectively. Panel B, high resolution map of TMEM154 regions targeted for PCR-amplification. PCR amplification primers are indicated with black arrowheads and listed in Table S3. Red lowercase letters above SNP positions are IUPAC/IUBMB ambiguity codes for nucleotides (r = a/g, y = c/t, m = a/c, k = g/t, s = c/g, w = a/t) [17] and indicate 12 sites affected by nonsynonymous substitutions. The red uppercase letters above SNP positions indicate the amino acid polymorphisms encoded at TMEM154 codons 4, 13, 14, 25, 31, 33, 35, 44, 70, 75, 82 and 102. Black lowercase letters below SNPs indicate nucleotide polymorphisms that resulted in synonymous substitutions. Panel C, the areas of circles for haplotypes 1 to 4 are proportional to the frequencies in the international panel of 75 ISGC sheep. The symbols are as follows: black circles, risk factors; white circle, non-risk factor; grey circles, risk factor status unknown; red circles, haplotypes known in U.S. sheep but not observed in the international panel of 75 ISGC sheep (risk factor status unknown); shaded square, TMEM154 haplotype predicted to have occurred but unobserved to date. Dashed grey line, haplotypes observed in wild sheep species but not domestic sheep. Haplotypes 13 and 14 were observed in one animal each and their location of origin is indicated.