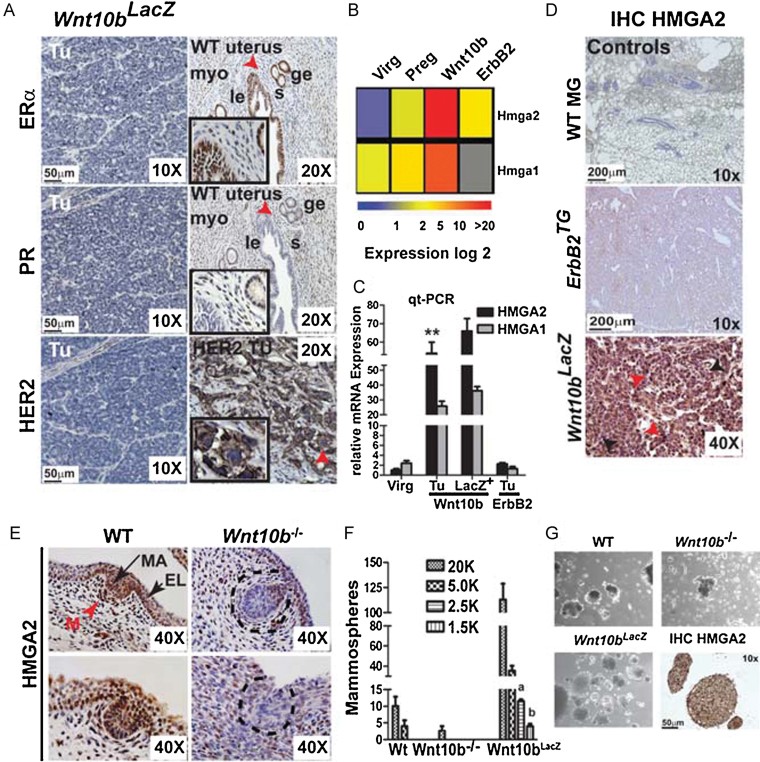
A. IHC for oestrogen receptor α (ERα), progesterone receptor (PR) and Her2 to validate the triple-negative phenotype of Wnt10bLacZ tumours. Wild-type (WT) uterus and Her2 tumour samples were used as positive controls. Highlighted are myoepithelial (myo), glandular epithelial (ge), luminal epithelial (le) and stromal cells (s).
B. Hierarchical clustering of microarray data of Hmga2 and Hmga1 expression in Wnt10b-driven tumours.
C. Validation of Hmga2 and Hmga1 expression in tumour cells (Tu), Lin−LacZ+ (LacZ+) tumour cells compared to wild-type virgin (Virg) and ErbB2TG tumour cells, as determined by qt-PCR. Error bars represent the means and the standard deviations from three independent experiments; **p-value = 0.007 versus Virg or ErbB2TG Tu samples (Student's t-test).
D. IHC of HMGA2 in WT mammary glands, and ErbB2TG and Wnt10bLacZ tumours. Red arrowheads highlight cells positive for HMGA2 and black arrowheads negative cells, respectively.
E. IHC of HMGA2 in WT and Wnt10b knockout (Wnt10b−/−) mammary placodes (mp) at embryonic Day 14.5 reveals loss of HMGA2 expression in Wnt10b−/− mp. Dashed black lines indicate Wnt10b−/− mp. Mesenchymal (M: red arrow head), epidermis layer (EL: short black arrow) and mammary anlagen (MA: long black arrow).
F, G. Mammosphere assays (MSA) of WT and Wnt10b−/− mammary gland cells and MMTV-Wnt10bLacZ primary tumour cells that were serially passaged (at Day 10 and 20) and analysed at Day 30. Secondary spheres were counted in sequential dilutions and imaged using a phase contrast microscope. Biological replicates (n = 3) and six technical replicates are shown. Wnt10bLacZ-derived MSAs analysed by IHC for HMGA2 is shown (10×) in lower right panel of G (bar, 50 µm). In F error bars represent the means and the standard deviations from three independent experiments; p values: a = 0.04, b = 0.03 versus wild-type or Wnt10b−/− samples (Student's t-test). p Values of <0.05 were considered to be statistically significant (C, F).