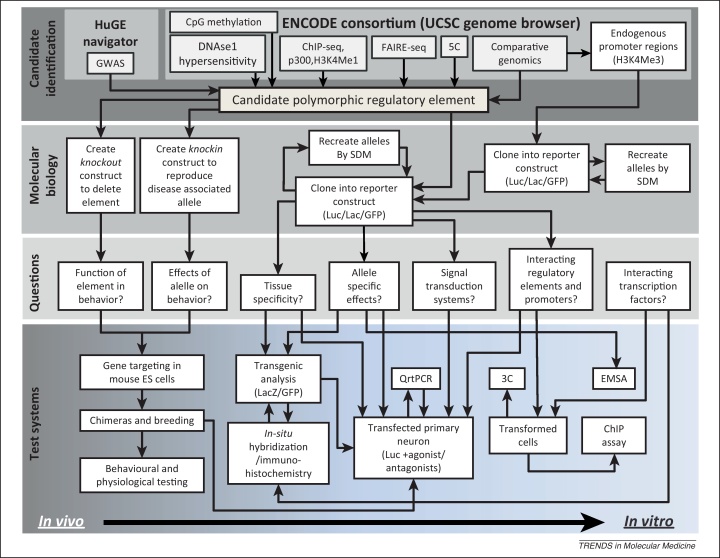
Figure 2.
A flow diagram describing the relationships between different technologies that can be used to identify and characterize cis-regulatory sequences (CRSs) and to determine the effects of polymorphisms on their qualitative and quantitative activities using a series of different in vivo, in vitro, and high-throughput technologies. The first row describes the technologies that can be used to identify CRSs (GWAS, genome-wide association analysis; ChIP-seq, chromatin immunoprecipitation sequencing; FAIRE-seq, formaldehyde-assisted identification of regulatory element sequencing; 5C, carbon copy chromatin conformation capture). The second row describes the genome and DNA manipulations required to test hypotheses relating to CRS activity and the effects of polymorphisms (SDM, site-directed mutation; Luc, luciferase; Lac, LacZ gene encoding β-galactosidase; GFP, green fluorescent protein). The third row summarizes many of the different questions relevant to the understanding of the function of CRSs and the effects of SNPs on their activity. The last row summarizes several different paradigms that can be used to address the questions posed in the third row (ES, embryonic stem cell; QrtPCR, quantitative reverse transcriptase polymerase chain reaction; 3C, chromatin conformation capture; EMSA, electrophoretic mobility shift assay; ChIP, chromatin immunoprecipitation). This flow diagram is not exhaustive and does not include technologies that allow analysis of epigenetic modification.