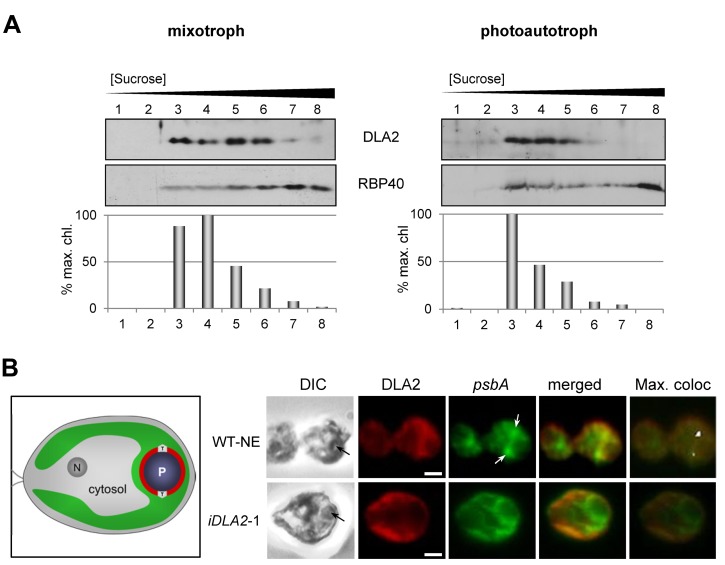
Figure 8. DLA2-dependent localization of the psbA mRNA to the PSII biogenesis region of the chloroplast (T-zone) and CTMs.
(A) Membranes from cells from either mixotrophic (left panel) or photoautotrophic (right panel) conditions were fractionated by isopycnic sucrose gradient ultracentrifugation, and fractions were analyzed by immunoblot for DLA2 and a marker protein for CTM (RBP40). As a marker for thylakoid membranes, chlorophyll concentration is graphed as the percentage of the maximum value. (B) Cells of either the control strain transformed with the empty vector (WT-NE) or DLA2-deficient (iDLA2-1) were grown under mixotrophic conditions, IF-stained with the α-DLA2-antiserum (red), and FISH-probed for the psbA mRNA (green). Black arrows in differential interference contrast (DIC) images at the left indicate the position of the pyrenoid. White arrows indicate where the psbA FISH signal is localized in T-zones. Pixels with the strongest signals obtained by the program Colocalization Finder (ImageJ) were labeled white (max. confocal). Bars, 1 µm. An illustration of a C. reinhardtii cell shows the nucleus (N), cytosol, and chloroplast (green) with the T-zone (T) and pyrenoid (P).