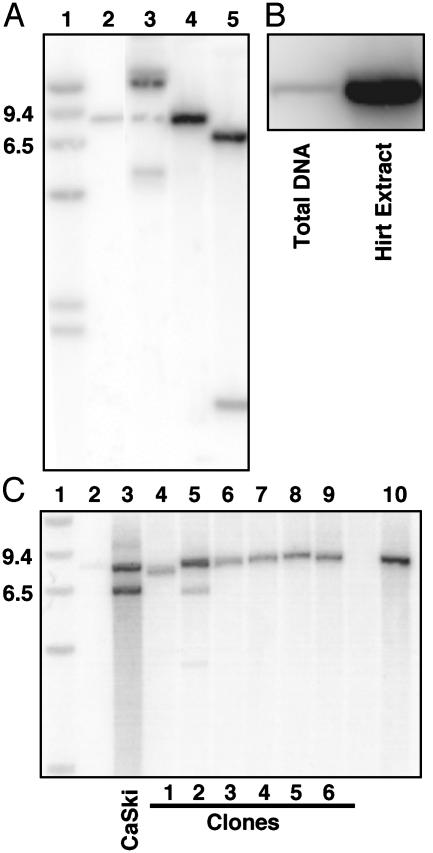
Fig. 3.
Southern blot hybridization of total and Hirt supernatant DNA extracted from cervical cells containing HPV16. (A) Establishment of episomal HPV16 genomes in cervical cells. Southern blot of restriction enzyme digests of total cellular DNA harvested 44 days after infection with Ad/HPV16/eGFP and Ad/Cre. Lane 1, 9.4- and 6.5-kb molecular weight markers. Lane 2, 3 pg of BamHI linearized HPV16 DNA. Lanes 3-5, total DNA from HPV16-containing cervical cells digested with PacI (no restriction sites in HPV16), BamHI (one restriction site in HPV16), and SphI plus BamHI (one site each at nucleotides 7,465 and 6,152), respectively. The majority of the uncut DNA form in lane 3 is present as an open circular DNA form, which is attributed to the DNA isolation technique because the amount of each DNA form varied among isolations (data not shown). (B) DNA from cervical cells isolated as total DNA or as a Hirt supernatant, which enriches episomal DNA. Equal quantities of DNA were digested with BamHI and loaded in each lane. (C) Assay for integration of HPV16 in isolated cell clones. Lanes 2 and 10 show 3 and 30 pg of linearized HPV16 DNA, respectively, as described in A. Lane 3-9, BamHI restriction enzyme digests of total cellular DNA isolated from CaSki cells (a positive control for integrated HPV16 DNA) and compared with DNA from single-cell clones 1-6.