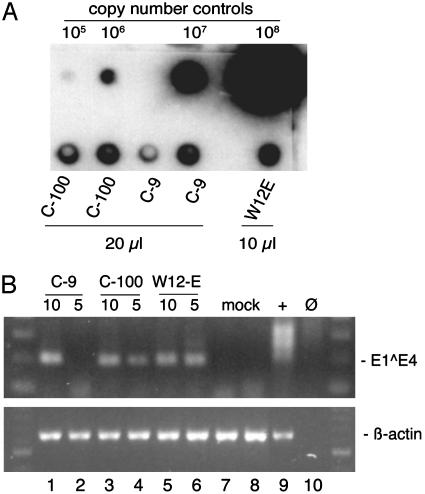
Fig. 6.
Quantification of virion production and analysis of reinfection in HaCaT cells. (A) Virions were extracted from organotypic tissues and analyzed by dot blot hybridization. Copy number control HPV16 DNA from cloned genomes ranging from 105 to 108 vge and 10 or 20 μl of virion preparations were denatured with sodium hydroxide, transferred to a nylon membrane, and then hybridized with a radioactively labeled HPV16 genome probe and exposed to a phosphorimage screen for quantification. (B) HPV16 virions isolated from a cloned line (C-9), a total cell population (C-100), and W12-E cells grown as raft tissues were used to infect HaCaT cells. The HaCaT cells were harvested 2 days after infection. Total RNAs (3 μg) were reverse-transcribed. (Upper) RNAs were analyzed from W12-E cells known to contain E1^E4 message (+, lane 9); mock-infected HaCaT cells (mock, lanes 7-8); HaCaT cells infected with a multiplicity of infection corresponding to ≈10 and 5 vge per cell, respectively, from C-9 (lanes 1-2); C-100 (lanes 3-4); and W12-E (lanes 5-6). No RNA input (Ø, lane 10) served as a negative amplification control. (Lower) β-actin primers were used to detect a 641-bp amplimer derived from spliced β-actin RNA in one round of PCR. The input RNA corresponded to 2.7 μg for HPV16 E1^E4 and 0.3 μg for β-actin. Molecular size standards (100-bp ladder) are shown in the lateral lanes.