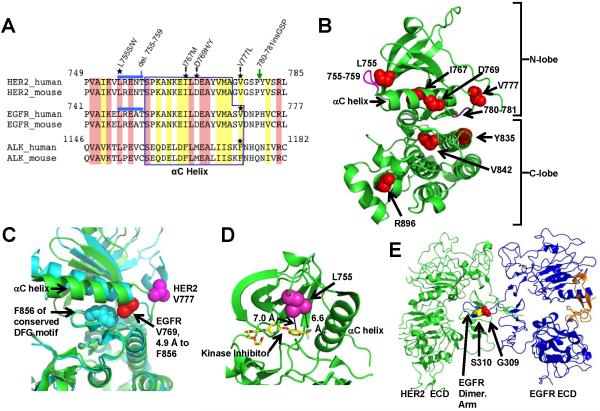
Figure 2.
Protein structure visualizations of the HER2 somatic mutations. A, Multi-sequence alignment of HER2, EGFR, and ALK tyrosine kinases. Location of somatic mutations is marked with stars. Blue line and green arrow indicate del.755-759 and P780ins, respectively. Identical residues are shaded pink and similar residues yellow. B, HER2 kinase domain structure (PDB: 3PP0) showing the locations of point mutations (red spheres) and deletions/insertions (magenta). C, HER2 V777 and EGFR V769 are visualized in the active conformation of the EGFR kinase domain (cyan, PDB: 2GS2) and the inhibitor bound conformation of HER2 (green, PDB: 3PP0). D, Proximity of the HER2 L755 to the tyrosine kinase inhibitor SYR127063 is shown. SYR127063 has the same core chemical structure as lapatinib (23). E, Alignment of HER2-EGFR extracellular domain structures PDB: 1N8Y (HER2) and PDB: 1IVO (EGFR).