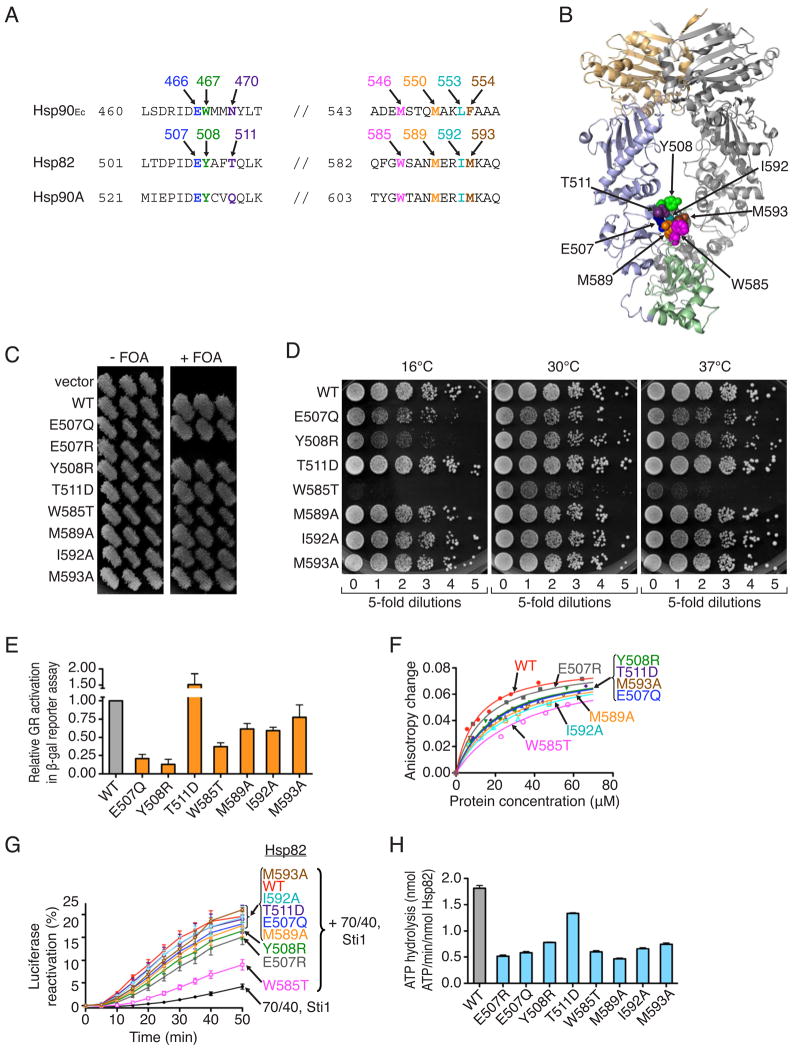
Figure 5. Several amino acids residues important for Hsp90Ec function are also important for yeast Hsp82 function.
(A) Alignment of E. coli Hsp90Ec, S. cerevisiae Hsp82 and human Hsp90A showing mutated residues.
(B) Model of the Hsp82 dimer made from the X-ray structure of Hsp82 in the closed conformation (pdb: 2cg9) visualized using PYMOL (www.pymol.org). In one protomer, the N-terminal domain is colored tan, the middle domain is light blue and the C-terminal domain is green. Residues that were mutated are represented as CPK models.
(C) Ability of Hsp82 mutants to support growth was assessed via a plasmid shuffle assay as described in Supplemental Information. S. cerevisiae G612 lacks chromosomal copies of HSC82 and HSP82 and is supported by wild type HSP82 on a URA3 plasmid. Strains with HSP82 alleles on LEU2 plasmids that support viability grow on FOA, which prevents growth of cells carrying the URA3-based HSP82 plasmid. Three individual transformants are shown.
(D) Overnight cultures of strains expressing Hsp82 wild type or mutants that supported growth were diluted to equal density, and aliquots of five-fold serial dilutions were spotted onto YPAD plates and incubated for three days at the indicated temperatures. The cells expressing the Hsp82 defective mutants looked normal by microscopy at the permissive temperature.
(E) Glucocorticoid receptor maturation. Strains expressing Hsp82 wild type or mutants that supported growth were transformed with plasmids encoding GR and a downstream LacZ reporter (Louvion et al., 1996). After growth in the presence of deoxycorticosteroid, β-galactosidase activity was measured. Values are averages of three independent measurements reported as relative to wild type. Error bars indicate standard deviations.
(F) Binding of Hsp82 wild type or mutants to IAEDANS-labeled Δ131Δ was measured by fluorescence anisotropy. Binding curves are the average of two independent measurements.
(G) Reactivation of heat-denatured luciferase by Hsp82 wild type or mutants in conjunction with Hsp70 and Ydj1 (70/40), and Sti1 was measured over time. Data from three replicates are presented as mean ± SEM.
(H) The ATPase activity of Hsp82 wild type or mutants was measured. Data from three replicates are presented as mean ± SEM.
See also Figure S5.