Abstract
The oxidation-reduction potentials of electron transfer proteins determine the driving forces for their electron transfer reactions. While the type of redox site determines the intrinsic energy required to add or remove an electron, the electrostatic interaction energy between the redox site and its surrounding environment can greatly shift the redox potentials. Here, a method for calculating the reduction potential versus the standard hydrogen electrode, E°, of a metalloprotein using a combination of density functional theory and continuum electrostatics is presented. This work focuses on the methodology for the continuum electrostatics calculations, including various factors that may affect the accuracy. The calculations are demonstrated using crystal structures of six homologous HiPIPs, which give E° that are in excellent agreement with experimental results.
Keywords: Density functional theory, Poisson-Boltzmann continuum electrostatics, reduction potentials, iron-sulfur proteins, metalloproteins
Introduction
Electron transfer reactions play an important role in many bioenergetic processes.1 Most biological electron carriers are proteins, which have redox sites that undergo oxidation-reduction reactions. The reduction potentials of these proteins are essential functional characteristics because they determine the driving forces for electron transfers, with the favorable direction toward the redox site of higher reduction potential. At the lower end of biological range is the splitting of water into H+ and O2 at an oxidation potential of −0.8 V, which occurs in photosynthesis, while the higher end is formation of water at a reduction potential of +0.8 V, which occurs in respiration1. However, proteins contain only a limited number of different types of redox sites to cover this range, so electron transfer pathways rely on the protein environment to tune the reduction potential to the appropriate value. For instance, multiple [4Fe-4S] redox sites are used in the electron transport chains in mitochondrial respiratory complex I2 and chloroplastic photosystem I3; the former has six [4Fe-4S] redox sites with reduction potentials spanning a range between −0.3 and −0.1 V4 and the latter has three [4Fe-4S] redox sites with reduction potentials spanning a range between −0.55 and −0.59 V5,6.
The effects of the protein environment on the reduction potential can be understood by considering electron transfer as the reduction reaction for a redox site A
| (1) |
where m is the net charge of the redox site in the oxidized state. The standard reduction potential against the standard hydrogen electrode, E°, for A can be related to the free energy ΔG° of the above reduction reaction using the Nernst equation
| (2) |
where n is the number of electrons being transferred and F is the Faraday constant. In the second equality, ΔGin is the change in the inner sphere energy upon reducing the redox site, ΔGout, is the change in the interaction energy of the redox site upon reduction with the outer sphere environment, and ΔGSHE is the free energy for the removal of an electron from the standard hydrogen electrode (SHE). For a simple ion in solution, the ion is the inner sphere and the solvent is the outer sphere while for a small molecule in solution, the redox active part of the molecule may be defined as the inner sphere and the outer sphere may include parts of the ligands not directly bonded to the metals in addition to the solvent. For a redox site in a metalloprotein, the inner sphere may be defined as the metal and its ligands so that the outer sphere consists of the protein and surrounding solvent. Thus, to the extent that the protein does not affect the electronic structure of the redox site, the outer sphere contribution is mainly responsible for differences in reduction potentials between different proteins with the same redox site.
Computational studies have played an important role in understanding reduction potentials of metalloproteins. Many earlier studies of proteins have assumed that ΔGin for a given redox site is constant and thus have focused on understanding relative values of E° between different proteins with the same redox site rather than E°. For instance, Warshel and co-workers have calculated relative E° for cytochrome c and iron-sulfur proteins via the protein-dipoles-Langevin dipoles (PDLD) method7,8 and Gunner and Honig have calculated relative E° of heme proteins using Poisson-Boltzmann (PB) continuum electrostatics9,10. In addition, Ichiye and coworkers have calculated differences in E° of as little as 0.05 V due to specific residues referred to as sequence determinants in iron-sulfur proteins11–15 using molecular mechanics combined with bioinformatics analysis. Although these earlier studies have provided much insight into reduction potentials of metalloproteins, it is difficult to evaluate different methods for calculating the protein contribution since deviations from experiment can be attributed to the missing inner sphere contribution. Thus, calculations of reduction potentials versus the SHE for [4Fe-4S] proteins by Noodleman, Case, Bashford and co-workers using broken-symmetry (BS) density functional theory (DFT) with the BP86 functional to calculate ΔGin and PB electrostatics to calculate ΔGout are an important step forward16. However, more recent studies17 indicate that inner sphere redox energies calculated with the BP86 functional differ considerably from electrospray ionization - photoelectron spectroscopy (EI-PES) detachment energies of [4Fe-4S] cluster analogs measured by Wang and co-workers18–20.
Recently, our group has predicted E° vs. SHE for [4Fe-4S] proteins that are in excellent agreement with experiment using a simple additive approach we refer to as DFT+PB in which ΔGin is obtained by BS-DFT calculations of gas phase redox site analogs and ΔGout is obtained by PB calculations of the redox site in the protein using partial charges from the BS-DFT calculation21. This may be considered a first order approximation since the interaction between the perturbation of the electronic structure of the redox site is ignored; however, surprisingly good results are obtained. A key factor has been our calibration of the functionals and basis sets for the BS-DFT calculations by comparing the calculated detachment energies of redox site analogs in the gas phase against the EI-PES data of Wang and co-workers, with the best agreement using B3LYP and double-ζ basis sets with the appropriate polarization and diffuse functions17. Our BS-DFT calculations also show that the value of ΔGin for the iron-sulfur clusters is relatively independent of environment17, since environmental factors (such as hydrogen bonds to the redox site) affect the electron detachment energy mainly by electrostatic contributions rather than perturbations of the electronic structure, thus making the DFT+PB approach possible22. Our BS-DFT calculations further show that the ligand dihedral conformations may tune the energies by ~100 mV, but this falls within ΔGin23. However, the protein environment may tend to polarize the partial charges of the redox site so a strategy similar to that of the CHARMM force field is adapted by using the 6–31G** basis set, which tends to overestimate polarization.24 Another key factor in the DFT+PB method is the accuracy of the calculation of ΔGout by PB electrostatic methods; for instance, the deviation in the calculated E° from experiment due to the resolution of the crystal structure used in the calculation has been previously estimated as < ~−0.04R to 0.09R eV, where R is the resolution21. Finally, the value of ΔGSHE comes from the half-cell potential for the SHE. IUPAC recognizes ΔGSHE as 4.44 eV, which is determined as summation of the free energy for the atomization of hydrogen, the ionization free energy of a hydrogen atom, and the solvation free energy of H+25. Another commonly accepted value is 4.43 eV determined by Reiss and Heller26 using a thermodynamic cycle for the removal of an electron from a saturated Pt electrode, although the determination of the solvation free energy of a proton remains an issue27,28.
The DFT+PB methods are demonstrated here for the [4Fe-4S] proteins; however, the methodology is being generalized to other metalloproteins for implemention of user-friendly redox potential calculations that have a reasonable estimate for both the inner and outer sphere contributions. To this end, standard parameters for partial charges and atomic radii of the protein including liganding side chains and standard values for the dielectric permittivity of the protein and ΔGSHE, are used so that the PB calculations can be made consistent. In addition, since the stumbling block for non-computational users is generally in the DFT calculations of the metal sites, which are CPU intensive and can be off by 1 eV in energy if the wrong functional is used,17 a library of ΔGin, partial charges, and atomic radii of the redox sites is being developed, in which the DFT approach has been calibrated against ES-PES data for analogs.
Here, the focus is on the methods used in the calculation of ΔGout for [4Fe-4S] proteins by PB continuum electrostatic methods. First, the procedure for obtaining ΔGout from quantities reported by typical PB equation solvers is described, including the proper reference states, treatment of atoms at the boundary, and techniques to reduce errors from the grid approximations. Next, the effects of the grid and factors concerning the protein structure are assessed. The effects of the assignment of the redox layers for the [4Fe-4S] sites, the oxidation state of the protein crystal, and the assignment of the nitrogen and oxygen of the carboxamides of asparagines and glutamines are examined. Finally, the effects of different basis sets on the calculation of E° and the overall accuracy with respect to experiment are examined. The procedure is demonstrated using six HiPIPs, but has shown to be accurate for two other types of ferredoxins and the nitrogenase iron-proteins as well21. Since these three types of [4Fe-4S] proteins have different folds with different exposures of the redox site, the methods are expected to be general for the [4Fe-4S] proteins. A discussion of generalizing the methods to other redox sites is included.
Methods
All protein crystal structures were obtained from the Protein Data Bank (PDB)29. Crystal structures of six HiPIP species were utilized: Rhodocyclus tenuis (Rt) [1ISU] at 1.50 Å resolution30, Rhodoferax fermentans (Rf) [1HLQ] at 1.45 Å resolution31, Ectothiorhodospira vacuolata (Ev) [1HPI] at 1.80 Å resolution32, Ectothiorhodospira halophila (Eh) [2HIP] at 2.50 Å resolution33, Thermochromatium tepidum (Tt) [3A39] at 0.72 Å34, [1IUA] at 0.80 Å35, [2FLA] at 0.95 Å (reduced), and [2AMS] at 1.40 Å resolution, and Chromatium vinosum (Cv) [1CKU] at 1.20 Å resolution36. For crystal structures containing more than one molecule in the asymmetric unit, the calculated energies were averaged over all structures with the exception of Eh; chain B from the Eh structures contained unusual Fe-S bond lengths so it was not utilized. Hydrogen atoms were added to the crystal structures with the HBUILD facility in CHARMM 35b1.37 Since inconsistencies in naming atoms in the PDB files can lead to large errors, the naming for the redox cluster, histidine residues, and terminal residues were corrected to match the parameter sets.
The ΔGin and partial charges of [Fe4S4(SCH3)4]n (n = 1-, 2-, 3-) are from BS-DFT calculations using the NWChem program package38 with the B3LYP exchange-correlation functional39,40 as previously described by Niu and Ichiye17. The Cβ-Sγ-Fe-Si (where Si is the inorganic sulfur on the opposite plane from the iron) dihedral angles, χ3, were ~60° in one redox layer and ~−60° in the other redox layer. Briefly, the geometries were optimized using the 6–31G**41 and DZVP242 basis sets with a fine integration grid of 140 radial shells and 974 angular points per shell (140,974) for Fe, (123,770) for S, (70,590) for C, and (60,590) for H. The vibrational analysis for the free energies and CHELPG43 electrostatic potential (ESP) charges were calculated at the level of the geometry optimization. Single point energies with added sp-type diffuse functions on the sulfurs of 6–31G**41,42 were calculated from the 6–31G** geometries, and are referred to as 6–31(++)SG**//6–31G** basis set calculations. Most of the calculations here used ΔGin = −0.232 (3.453) eV for the 1-/2- (2-/3-) redox couple at the B3LYP/6–31(++)SG**//B3LYP/6–31G** level of theory/basis set and partial charges at B3LYP/6–31G** level of theory/basis set. In addition, calculations using ΔGin = −0.371 (3.369) eV for the 1-/2- (2-/3-) couple and partial charges both at the B3LYP/DZVP2 level/basis set were examined. ΔGSHE = 4.43 eV26 was utilized. All partial charges were averaged such that equivalent atoms on each redox layer had the same charge. In addition, to account for the difficulty in assigning charges for buried atoms in the methyl groups, all hydrogen atoms were assigned a charge of 0.09 e and the charge on the carbon to which they were attached was assigned a charge such that the net charge on the entire methyl group was maintained. The resulting partial charges are reported in Table S1 and S2 of the supplementary material.
The Poisson continuum electrostatic calculations were performed using APBS,44 a program for solving the PB equation. In addition, these calculations can now be performed using our implementation for redox calculations in CHARMMing,45 a web-based interface for the program CHARMM that now includes the APBS program37. Although the ionic concentration here is zero, the calculations are referred to as “PB” following common practice. Unless otherwise noted, calculations were performed in a 51.2 Å × 51.2 Å × 51.2 Å cubic grid with a grid spacing of 0.2 Å × 0.2 Å × 0.2 Å. Charges were mapped onto the dielectric grid with a cubic B-spline discretization and a multiple Debye-Hückel boundary condition was applied to all calculations. The protein in solution has three dielectric regions representing the redox site, the protein, and the solvent, with permittivities εc = 1, εp = 4, and εs = 78, respectively. The dielectric maps were constructed using the Connolly surfaces46 of the redox site and the protein with a probe radius of 1.4 Å and the boundary grid points were smoothed as the average of the original values of the grid point and its eight nearest neighbors.47 The three-region dielectric maps were constructed from two maps: one in which points within the redox site surface were assigned εc = 1 while those outside were assigned εp = 4 and another in which all points within the protein surface were assigned εp = 4 while those outside were assigned εs = 78. Using the DXMATH program, which installs with APBS, the final map was constructed by adding the dielectric maps together and subtracting εp = 4 from each point for the over-counting. The partial charges for the protein and the atomic radii for all protein atoms are taken from the CHARMM22 parameter set.24 For the redox site, the radius for iron is 1.07 Å48, the same radii for the cysteinyl ligands are used as equivalent protein cysteine atoms, and the radius for inorganic sulfur is chosen to be equivalent to the cysteinyl sulfur; i.e., 2.00 Å. The assignments of the mixed-valance layers for all HiPIPs were based on NMR experiments for CvHiPIP in which the first two Cys in the sequence bind the “top” layer and the second two Cys bind the “bottom” layer, with the top layer being the more oxidized layer49.
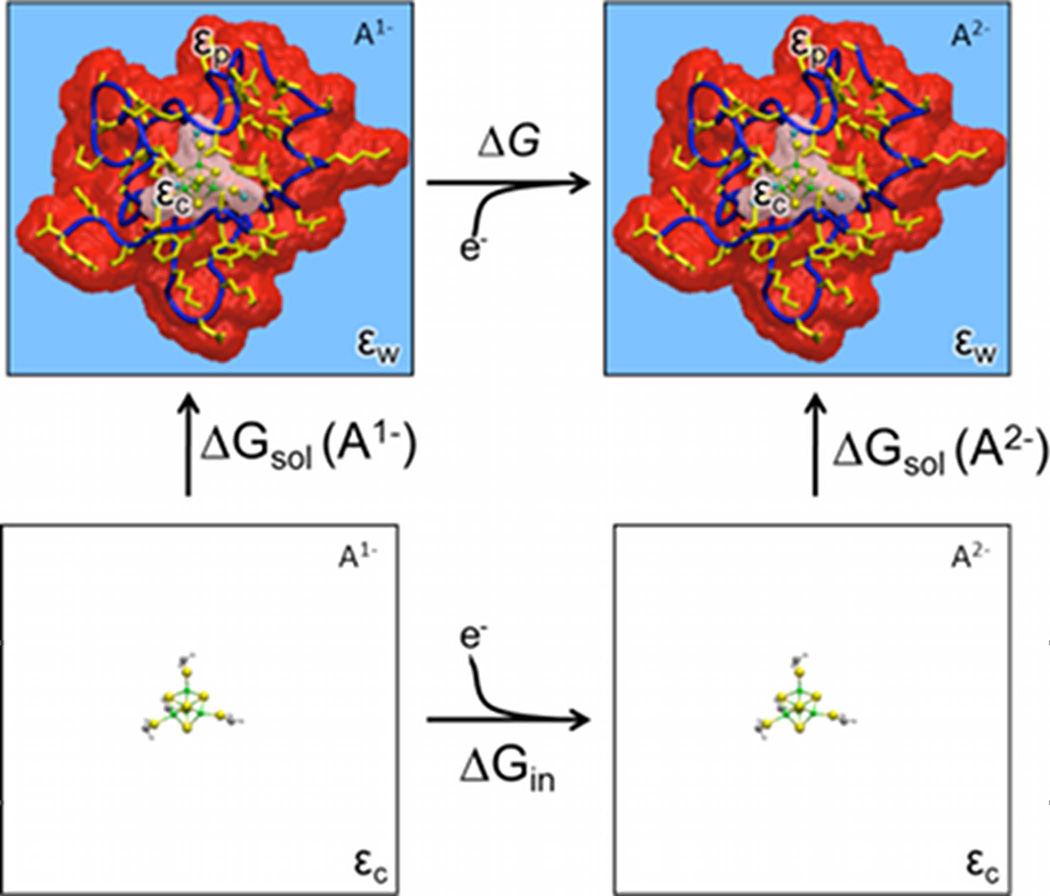
The absolute free energy ΔG for the reduction reaction of a protein (Eqn. 1) can be calculated using a thermodynamic cycle (Figure 1). Since the overall reduction can be separated into the reduction of the redox site in vacuum, and the solvation of the redox site in both the oxidized and reduced states, ΔGout is the difference in ΔsolG(A), the solvation energy of the redox site A, between the oxidized (m) and reduced (m−1) states
| (3) |
where ΔsolG(A) is the interaction energy of the redox site with both the surrounding protein and the solvent; i.e.,
| (4) |
If the same structure is used for the protein to calculate ΔsolG(Am) for the oxidized and reduced states, then the last term in Eqn. 4 cancels in Eqn. 3. In addition, for comparisons between different proteins, all structures are aligned by the redox site to reduce errors arising from extrapolation of the partial charges onto the grid. Redox site atoms are identical in the solvated (i.e. in the protein and solvent) and in the reference state (i.e. in vacuo); thus, here the thiol sulfurs in the reference state are terminated by methylene groups.
Figure 1.
Results
The calculation of E° vs. SHE using our DFT+PB method is demonstrated here. First, the effects of the grid and various factors of the protein crystal structures are examined. Next, the effects of the basis set used for calculation of the partial charges and ΔGout for the calculations of E° are examined. The state of redox site is referred to by the net charge n of [Fe4S4(SCH3)4]n.
Grid Dependence
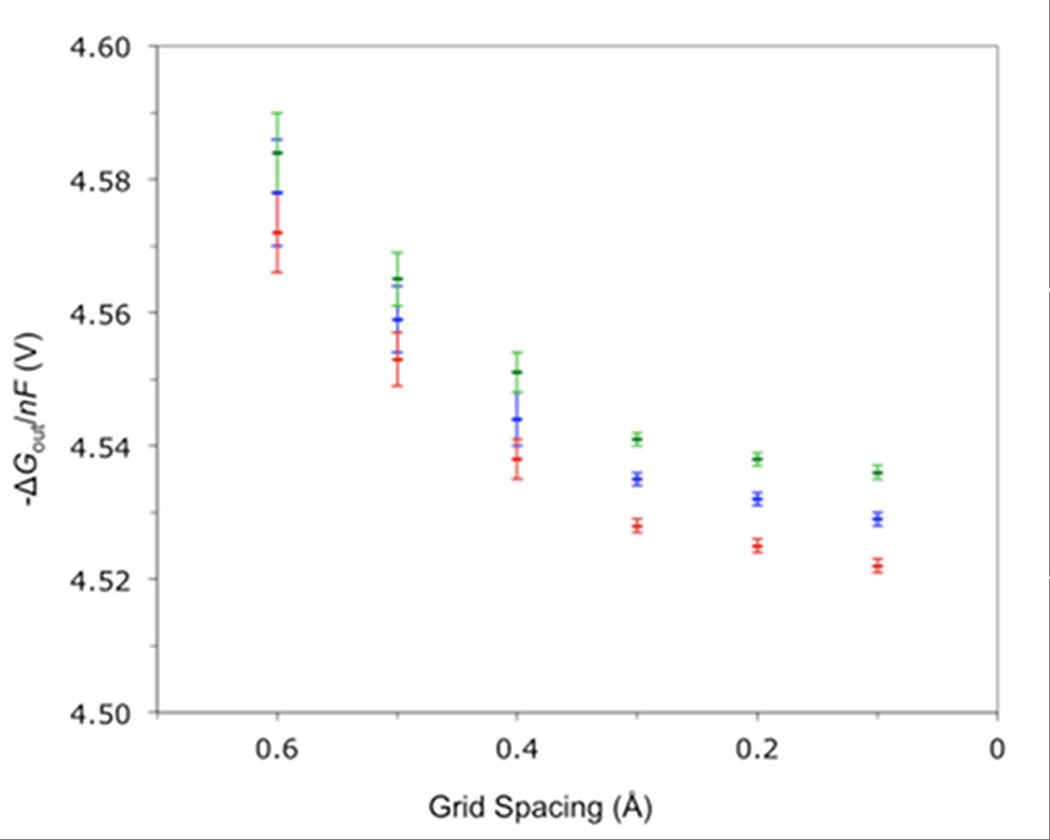
The dependence of ΔGout on grid spacing and the orientation of the molecule is demonstrated for 1-/2- couple of TtHiPIP in Figure 2. Calculations were performed at grid spacings between 0.6 and 0.1 Å, and, at each grid spacing, the protein and dielectric maps were rotated at 10° steps around the z-axis for 90°. All other parameters, such as the size of the calculation grid, were kept constant. The dependence on the grid spacing and the variation due to orientation began to decrease at grid spacings below 0.3 Å. Below this point, the continued decrease is most likely due to the method used to smooth the boundary between regions, which continues to become more sharp depending on the grid spacing. A single calculation of HiPIP using a 0.2 Å spacing took 17 minutes of CPU time and required 4 GB of memory, while at 0.1 Å spacing took 17.5 hours and required over 30 GB of memory (memory requirements are greater than 200 B per grid point). While the computational time at a 0.1 Å grid spacing is feasible for small proteins, the 0.2 Å grid spacing is deemed sufficient since the E° are only 2 to 3 mV larger, which is well within the error of the DFT+PB approximation, with much greater computational efficiency. All further calculations described are at the 0.2 Å spacing.
Figure 2.
Assignment of Redox Layers
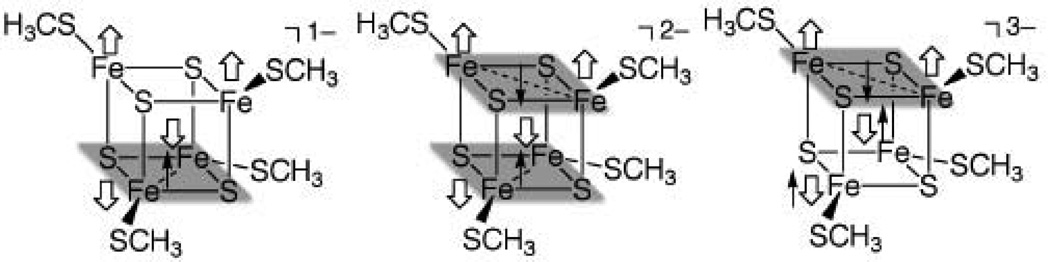
The cuboidal [4Fe-4S] cores are characterized by two redox layers, with each layer consisting of two irons and two sulfurs (Figure 3)50. The assignment of the layers may affect ΔGout because while the layers are equivalent in the 2- state, the oxidation or reduction from the 2- state occurs mainly in one layer, resulting in different partial charges in each layer. Each layer will be referred to as an n-m layer, where n and m are the formal charges on the two irons in the layers. In addition, when the irons should be formally 2+ and 3+, a minority spin becomes delocalized in the layer and each iron can be said to have a formal charge of 2.5+. Thus, in all states of the [4Fe-4S] redox site, each layer has the same partial charges for both irons, etc. (See supplementary material for partial charges). For the HiPIPs in the 1- state, the 3-3 layer can be assigned as having Cys43 and Cys46 ligands while the 2.5-2.5 layer can be assigned as having Cys61 and Cys75 ligands, according to 2D NMR studies of CvHiPIP,49 which is also consistent with the distortion of the [4Fe-4S] cubane found in our DFT studies.51 However, although it is reasonable to assume that the layer assignment is the same within a homologous set of proteins (i.e., the rest of the HiPIPs), the assignment may not be clear in other [4Fe-4S] proteins.
Figure 3.
Therefore, the effects of different assignments of the redox layers were tested in TtHiPIP for the 1-/2- couple, which leads to a ~0.03 V range in E° (Table 1). The largest deviations in E° occur when the irons bound to Cys43 and Cys75 are assigned to one layer and the irons bound to Cys46 and Cys61 are assigned to the other, since the Cys75 backbone amide interacts with the inorganic sulfur bridging the irons bound to Cys43 and Cys75, which leads to greater stabilization of negative charge on layers containing either of these two irons. Thus ΔsolG([Fe4S4(SCH3)4]1−) is more negative when the 2.5-2.5 layer is assigned as Cys43/75 and more positive when the 3-3 layer is assigned Cys43/75. However, as a whole, even incorrect assignment of layers will lead to relatively small errors since the range is only 30 mV.
Table 1.
Calculated ΔsolG([Fe4S4(SCH3)4]1−) and E° for different redox layer assignments for TtHiPIP. First line corresponds to the experimentally determined layer assignments.
| Top Layer | Bottom Layer | ΔsolG (V) | E° (V) |
|---|---|---|---|
| Cys 43, Cys 46* | Cys 61, Cys 75* | −2.428 | 0.334 |
| Cys 43, Cys 75 | Cys 46, Cys 61 | −2.444 | 0.318 |
| Cys 46, Cys 75 | Cys 43, Cys 61 | −2.428 | 0.335 |
| Cys 61, Cys 75 | Cys 43, Cys 46 | −2.429 | 0.334 |
| Cys 46, Cys 61 | Cys 43, Cys 75 | −2.411 | 0.351 |
| Cys 43, Cys 61 | Cys 46, Cys 75 | −2.428 | 0.335 |
| Exp | N/A | 0.321 |
Ref.53
Oxidation State of the Crystal Structure
The oxidation state of the protein crystal structure used for the calculation will also affect the calculated E°. Due to the difficulty of preventing oxidation of the crystal structure before the diffraction data is collected, most of the HiPIP structures have been solved at the 1- oxidation state. In the case of TtHiPIP, many crystal structures have been solved at different resolutions, with one structure solved in the 2- oxidation state (Table 2). The root mean-square deviation (RMSD) of the backbone atoms between the oxidized structures is relatively small and E° from the oxidized structures shows a range of less than 20 mV for the three structures from different resolutions. (Note: the 1IUA has a slightly larger RMSD due to differences remote from the redox site.) While the reduced structure has as a similar RMSD, E° from the reduced crystal structure is ~40 mV higher than the oxidized structures in both redox couples, which indicates that the reduced structure is more capable of stabilizing a negative charge at the redox site due to an increase in polarization of the protein around the redox site. For the biologically relevant 1-/2- redox couple of the HiPIPs, the E° from the oxidized structures are in better agreement with experiment. Although the 2-/3- couple cannot be seen in many HiPIPs, E° for this couple has been estimated as −0.64 V for Rhodopila globiformis HiPIP,52 and the E° from the reduced structure is in better agreement with this value. Thus, the correct oxidation state of the crystal appears to have a more significant effect on the calculated E°, although a reasonable estimate can be obtained even if the oxidation state is inconsistent with the redox couple of E°.
Table 2.
Calculated E° for TtHiPIP from crystal structures at different resolutions and oxidation states. The RMSD of the backbone atoms is calculated relative to the 0.72 Å oxidized crystal structure.
Nitrogen and Oxygen Assignment in Amides Groups
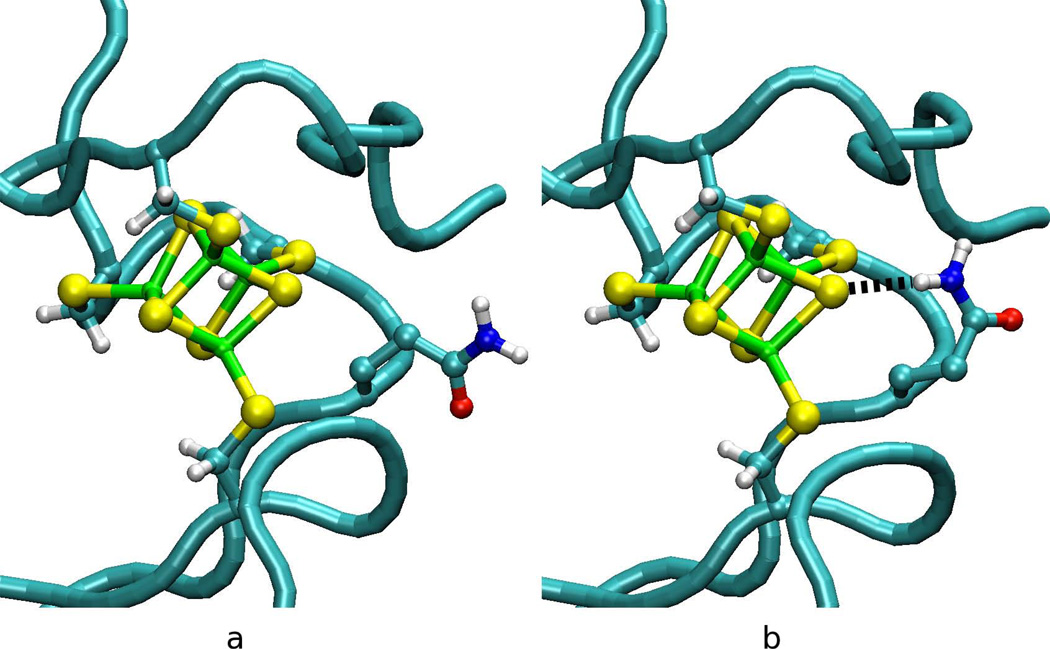
The effect on the calculated E° of the assignments of nitrogen and oxygen atoms in amide groups of Asn and Gln within 8 Å of the redox site was also examined for all of the HiPIPs (Table 3). Errors in assignnment has been estimated as 15%53,54 in x-ray crystallographic studies due to their similar electron density. Of the six structures, only RfHiPIP has no change in E° when the N and O are interchanged since the amine and carbonyl dipoles in Gln45 are perpendicular to the redox site and there is essentially no change in interaction energy, ΔEint, with the rest of the protein. The calculated E° for Cv, Tt, Eh and Ev HiPIP are slightly higher by 10 to 60 mV for the interchanged N and O while the calculated E° of Rt HiPIP is nearly 50 mV lower because the interchange breaks a hydrogen bond with the redox site in chain B of the asymmetric unit although it has little effect in chain A because Gln27 is oriented away from the redox site (Figure 4). However, the ΔEint is also considerably more favorable for the crystal assignment. so the in other cases, Asn or Gln side chains may be rotating as was seen previously in molecular dynamics simulations of rubrerythrin14 so that molecular dynamics simulations may be useful in examining calculated E° that disagree with experiment.
Table 3.
Calculated E° for the HiPIPs with the nitrogen and oxygen atoms of Asn or Gln in the crystal structure assignment and with nitrogen and oxygen exchanged. ΔEint, the change in interaction energy of the Asn or Gln with the rest of the proteins when the nitrogen and oxygen are interchanged, is also given.
| Species | Residue | ΔEint (kcal/mol) |
E° (V) | ||
|---|---|---|---|---|---|
| Crystal Assignment |
N/O Exchanged |
Experiment | |||
| Cv | Asn 45 | 28.4 | 0.384 | 0.394 | 0.361 |
| Tt | Asn 45 | 29.8 | 0.334 | 0.390 | 0.322 |
| Eh | Asn 33 | 25.2 | 0.104 | 0.138 | 0.111 |
| Ev | Asn 36 | 11.8 | 0.222 | 0.272 | 0.151 |
| Rf | Gln 45 | −1.5 | 0.350 | 0.349 | 0.333 |
| Rt | Gln 27 | 23.3 | 0.418 | 0.366 | 0.354 |
| Rt (a) | Gln 27 | 20.9 | 0.414 | 0.433 | |
| Rt (b) | Gln 27 | 25.9 | 0.423 | 0.299 | |
Figure 4.
Basis Set Dependence
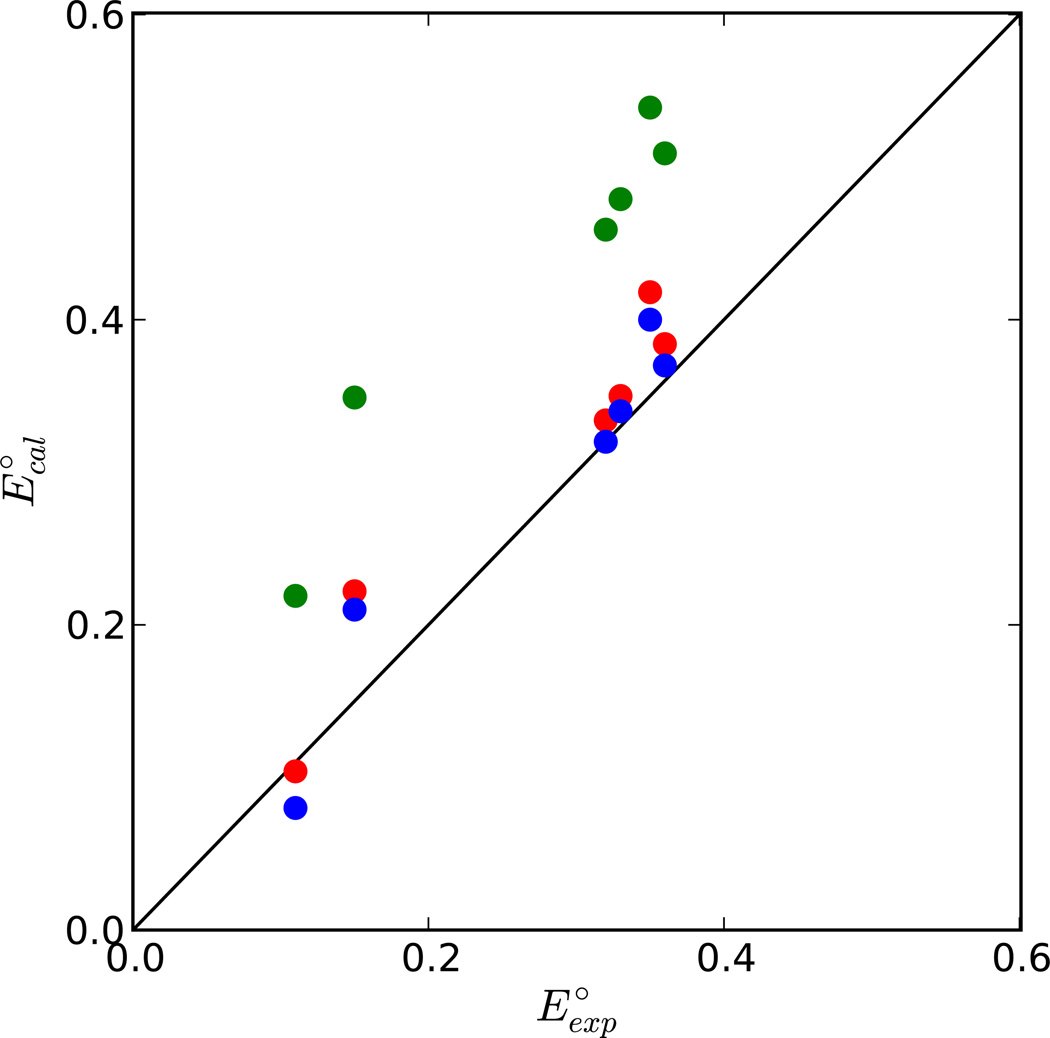
The dependence on the basis set used for calculation of the partial charges and ΔGin was examined by calculating E° for the 1-/2- couple of the HiPIPs and comparing with experiment (Figure 5). The partial charges at the B3LYP/6–31(++)SG**// B3LYP/6–31G** (i.e., the same level at which ΔGin was calculated) were not tested since they are only very slightly more polarized than those at B3LYP/6–31G** (i.e, the level of the geometry optimization, differing by only 0.03e on the Fe in the 2- state. E° using ΔGin calculated at B3LYP/6–31(++)SG**//B3LYP/6–31G** level/basis set combined with ΔGout using partial charges calculated at B3LYP/6–31G** level/basis set agree well with experiment. Since the DZVP2 charges are slight less polarized than the 6–31G** charges, differing by 0.15 e on the Fe in the 2- state, the calculated values of ΔGout for the six HiPIPs differ by only 0.015 ± 0.004 V. While this leads to slightly better agreement with experiment with ΔGin calculated at B3LYP/6–31(++)SG**//B3LYP/6–31G**, using ΔGin calculated at B3LYP/DZVP2 leads to much worse agreement with experiment. Moreover, other preliminary studies indicate that the slight overestimation by ΔGin at B3LYP/6–31(++)SG**//B3LYP/6–31G** level/basis set combined with ΔGout using partial charges at B3LYP/6–31G** level/basis set comes from the ligand dihedral angle dependence in the calculation of ΔGin.
Figure 5.
Discussion
While the methods are shown here for the [4Fe-4S] proteins, they will be generalized to other metalloproteins for implementation into user-friendly redox potential calculations in CHARMMing, as mentioned in the introduction. Of course, the PB calculations are highly dependent on the radii of the redox site atoms (although relatively insensitive to the metal radii since they are buried by their ligands) or alternatively to the choice of dielectric for the redox site so that the choices here may account for errors due to choice of ΔGSHE,28 polarization effects, etc. However, since the main goal is to provide two reasonable reference points, namely in the inner and outer sphere contributions, a best practices for each type of redox site can be established. Thus, the methods for each type of redox site are being tested in multiple proteins and in solvated analogs.
Conclusions
The DFT+PB method for calculating reduction potentials relative to the standard hydrogen electrode is presented in which the inner-sphere contribution is calculated by DFT as the change in energy of the relevant gas phase analog upon reduction while the outer sphere contribution is calculated by PB continuum electrostatics as the change in solvation energy of the redox site upon reduction. The calculated reduction potentials for [4Fe-4S] proteins are in excellent agreement with experimental values. Although the perturbations by the protein to the electronic structure of the redox site are neglected, previous calculations have shown that these are small and that the use of hybrid functionals and full-core basis sets are much more important in the redox energetics of the redox site. The influence of the protein is mostly electrostatic, and so using ESP charges for the different redox states to calculate the outer sphere contribution is sufficient. Moreover, this method allows deviations from experiment to be explored separately, as both perturbations to the electronic structure of the redox site and classical effects within the protein.
Several factors can affect the accuracy of the outer sphere continuum calculation. For instance, a grid spacing of 0.2 Å gives good results with reasonable computational times and memory requirements but larger spacing is not recommended. Although the results were relatively insensitive to the assignment of the redox layers, specific interactions may cause greater sensitivity. Also, the assignment of the nitrogen and oxygen atom of Asn and Gln that are close to the redox site may affect the E°; however, the assignments found in the crystal structures are reasonable in most cases. In addition, the oxidation state of the protein in the crystal structure appears to be important, possibly leading to errors of ~40 mV. Importantly, the results are relatively insensitive to the partial charges, while they are much more sensitive to the proper value of the inner sphere contribution. Thus, using ΔGin at the B3LYP/6–31(++)SG**//B3LYP/6–31G** level/basis set and ΔGout from partial charges at the B3LYP/6–31G** level/basis set is considered the most accurate and consistent combination for the [4Fe-4S] proteins.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by a grant from the National Institutes of Health (GM045303) and by the Intramural Research Program of the National Institutes of Health, National Heart, Lung, and Blood Institute in the Laboratory of Computational Biology (Z99-TW999999-03). The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of the U.S. Government. The continuum electrostatic calculations were performed on computers funded through the William G. McGowan Foundation and Georgetown University. The DFT calculations were performed at the EMSL, a national user facility sponsored by the U.S. DOE’s Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory, operated for DOE by Battelle, under the grant EMSL38793 and ST39962.
Reference
- 1.Lehninger A, Nelson DL, Cox MM. Lehninger Principles of Biochemistry. 4th ed. New York, NY: W.H. Freeman & Co.; 2003. [Google Scholar]
- 2.Sazanov LA. Biochemistry. 2007;46 doi: 10.1021/bi602508x. [DOI] [PubMed] [Google Scholar]
- 3.Romberger S, Golbeck J. Photosynth. Res. 2010;104:333. doi: 10.1007/s11120-010-9543-y. [DOI] [PubMed] [Google Scholar]
- 4.Reda T, Barker CD, Hirst J. Biochemistry. 2008;47:8885. doi: 10.1021/bi800437g. [DOI] [PubMed] [Google Scholar]
- 5.Ke B, Hansen RE, Beinert H. Proc. Natl. Acad. Sci. U. S. A. 1973;70:2941. doi: 10.1073/pnas.70.10.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heathcote P, Williams-Smith DL, Sihra CK, Evans MCW. Biochim. Biophys. Acta Bioenerg. 1978;503:333. doi: 10.1016/0005-2728(78)90192-5. [DOI] [PubMed] [Google Scholar]
- 7.Churg AK, Warshel A. Biochemistry. 1986;25:1675. doi: 10.1021/bi00355a035. [DOI] [PubMed] [Google Scholar]
- 8.Stephens PJ, Jollie DR, Warshel A. Chem. Rev. (Washington, DC, U. S.) 1996;96:2491. doi: 10.1021/cr950045w. [DOI] [PubMed] [Google Scholar]
- 9.Gunner MR, Honig B. Proc. Natl. Acad. Sci. U. S. A. 1991;88:9151. doi: 10.1073/pnas.88.20.9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao J, Hauser K, Gunner M. Biochemistry. 2003;42:9829. doi: 10.1021/bi027288k. [DOI] [PubMed] [Google Scholar]
- 11.Swartz P, Beck B, Ichiye T. Biophys. J. 1996;71:2958. doi: 10.1016/S0006-3495(96)79533-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ergenekan CE, Thomas D, Fischer JT, Tan ML, Eidsness MK, Kang CH, Ichiye T. Biophys. J. 2003;85:2818. doi: 10.1016/S0006-3495(03)74705-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eidsness MK, Burden AE, Richie KA, Kurtz DMJ, Scott RA, Smith ET, Ichiye T, Beard B, Min T, Kang C. Biochemistry. 1999;38:14803. doi: 10.1021/bi991661f. [DOI] [PubMed] [Google Scholar]
- 14.Luo Y, Ergenekan C, Fischer J, Tan M, Ichiye T. Biophys. J. 2010;98:560. doi: 10.1016/j.bpj.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beck BW, Xie Q, Ichiye T. Biophys. J. 2001;81:601. doi: 10.1016/s0006-3495(01)75726-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torres RA, Lovell T, Noodleman L, Case DA. J. Am. Chem. Soc. 2003;125:1923. doi: 10.1021/ja0211104. [DOI] [PubMed] [Google Scholar]
- 17.Niu S, Ichiye T. Mol. Simul. 2011;37:572. [Google Scholar]
- 18.Zhai H, Yang X, Fu Y, Wang X, Wang L. J. Am. Chem. Soc. 2004;126:8413. doi: 10.1021/ja0498437. [DOI] [PubMed] [Google Scholar]
- 19.Fu Y-J, Yang X, Wang X, Wang LS. Inorg. Chem. 2004;43:3647. doi: 10.1021/ic0495261. [DOI] [PubMed] [Google Scholar]
- 20.Wang XB, Wang LS. J. Phys. Chem. 2000;112:6959. [Google Scholar]
- 21.Perrin BS, Jr, Ichiye T. Proteins: Struct. Funct. Bioinf. 2010;78:2798. doi: 10.1002/prot.22794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, Niu S, Ichiye T, Wang L-S. J. Am. Chem. Soc. 2004;126:15790. doi: 10.1021/ja045709a. [DOI] [PubMed] [Google Scholar]
- 23.Niu S, Ichiye T. J. Am. Chem. Soc. 2009;131:5724. doi: 10.1021/ja900406j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. J. Phys. Chem. B. 1998;102:3586. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 25.Trasatti S. Pure Appl. Chem. 1986;58:955. [Google Scholar]
- 26.Reiss H, Heller A. J. Phys. Chem. 1985;89:4207. [Google Scholar]
- 27.Kelly CP, Cramer CJ, Truhlar DG. J. Phys. Chem. B. 2007;111:408. doi: 10.1021/jp065403l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamoureux G, Roux B. J. Phys. Chem. B. 2006;110:3308. doi: 10.1021/jp056043p. [DOI] [PubMed] [Google Scholar]
- 29.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rayment I, Wesenberg G, Meyer TE, Cusanovich MA, Holden HM. J. Mol. Biol. 1992;228:672. doi: 10.1016/0022-2836(92)90849-f. [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez A, Benini S, Ciurli S. Acta Crystallogr. Sect. D: Biol. Crystallogr. 2003;59:1582. doi: 10.1107/s0907444903014604. [DOI] [PubMed] [Google Scholar]
- 32.Benning MM, Meyer TE, Rayment I, Holden HM. Biochemistry. 1994;33:2476. doi: 10.1021/bi00175a016. [DOI] [PubMed] [Google Scholar]
- 33.Breiter DR, Meyer TE, Rayment I, Holden HM. J. Biol. Chem. 1991;266:18660. doi: 10.2210/pdb2hip/pdb. [DOI] [PubMed] [Google Scholar]
- 34.Takeda K, Kusumoto K, Hirano Y, Miki K. J. Struct. Biol. 2010;169:135. doi: 10.1016/j.jsb.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 35.Liu L, Nogi T, Kobayashi M, Nozawa T, Miki K. Acta Crystallogr. Sect. D: Biol. Crystallogr. 2002;D58:1085. doi: 10.1107/s0907444902006261. [DOI] [PubMed] [Google Scholar]
- 36.Carter CW, Kraut J, Freer ST, Alden RA. J. Biol. Chem. 1974;249:6339. [PubMed] [Google Scholar]
- 37.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J. Comput. Chem. 1983;4:187. [Google Scholar]
- 38.Valiev M, Bylaska EJ, Govind N, Kowalski K, Straatsma TP, Van Dam HJJ, Wang D, Nieplocha J, Apra E, Windus TL, de Jong W. Comput. Phys. Commun. 2010;181:1477. [Google Scholar]
- 39.Becke AD. J. Chem. Phys. 1993;98:5648. [Google Scholar]
- 40.Lee C, Yang W, Parr RG. Phys. Rev. B. 1988;37:785. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 41.Hehre WJ, Radam L, Schleyer PvR, Pople J. Ab initio molecular orbital theory. New York: Wiley-Interscience; 1986. [Google Scholar]
- 42.Godbout N, Salahub DR, Andzelm J, Wimmer E. Can. J. Chem. 1992;70:560. [Google Scholar]
- 43.Breneman CM, Wiberg KB. Journal of Comput. Chem. 1990;11:361. [Google Scholar]
- 44.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10037. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller BT, Singh RP, Klauda JB, Hodoscek M, Brooks BR, Woodcock HL., III J. Chem. Inf. Model. 2008;48:1920. doi: 10.1021/ci800133b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Connolly ML. Science. 1983;221:709. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- 47.Bruccoleri RE, Novotny JR, Davis ME, Sharp K. J. Comput. Chem. 1997;18:268. doi: 10.1006/jmbi.1997.0961. [DOI] [PubMed] [Google Scholar]
- 48.Jafri JA, Logan J, Newton M. Isr. J. Chem. 1980;19:340. [Google Scholar]
- 49.Bertini I, Capozzi F, Ciurli S, Luchina C, Messori L, Piccioli M. J. Am. Chem. Soc. 1992;114:3332. [Google Scholar]
- 50.Mouesca JM, Chen JL, Noodleman L, Bashford D, Case DA. J. Am. Chem. Soc. 1994;116:11898. [Google Scholar]
- 51.Niu S, Ichiye T. J. Phys. Chem. A. 2009;113:5710. doi: 10.1021/jp900402y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heering HA, Bulsink YBM, Hagen WR, Meyer TE. Eur. J. Biochem. 1995;232:811. [PubMed] [Google Scholar]
- 53.Donald IKM, Thornton JM. Protein Eng. 1994;8:217. [Google Scholar]
- 54.Word JM, Lovell SC, Richardson JS, Richardso DC. J. Mol. Biol. 1999;285:1735. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 55.Moulis J, Scherrer N, Gagnon J, Forest E, Petillot Y, Garcia D. Arch. Biochem. Biophys. 1993;305:186. doi: 10.1006/abbi.1993.1409. [DOI] [PubMed] [Google Scholar]
- 56.Luchinat C, Capozzi F, Borsari M, Battistuzzi G, Sola M. Biochem. Biophys. Res. Commun. 1994;203:436. doi: 10.1006/bbrc.1994.2201. [DOI] [PubMed] [Google Scholar]
- 57.Hochkoeppler A, Ciurli S, Venturoli G, Zannoni D. FEBS Lett. 1995;357:70. doi: 10.1016/0014-5793(94)01334-w. [DOI] [PubMed] [Google Scholar]
- 58.Heering HA, Bulsink YBM, Hagen WR, Mayer TE. Biochemistry. 1995;34:14675. doi: 10.1021/bi00045a008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.