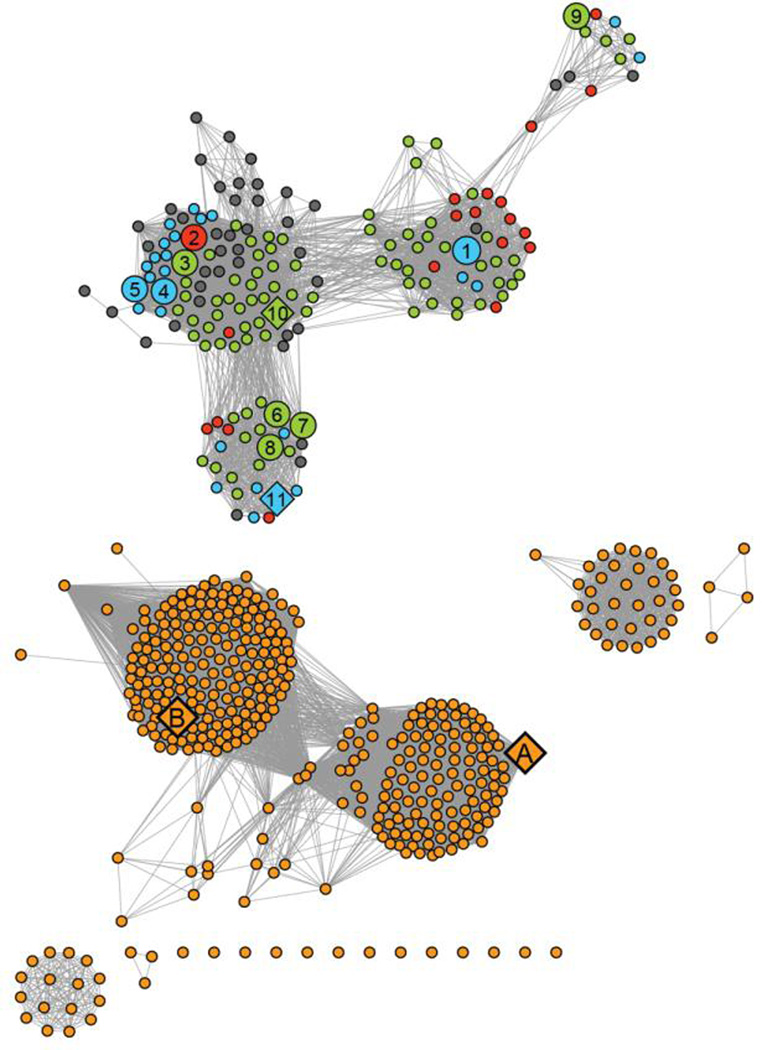
Figure 2.
Sequence similarity network of proteins in cog 1387 at an E-value of 10−20 created using Cytoscape (http://www.cytoscape.org). Each node (sphere) represents a single sequence and each edge (line) represents the pairwise connection between two sequences with the most significant BLAST E-value (better than 10−20). Lengths of edges are not significant, except for tightly clustered groups, which are more closely related than sequences with only a few connections. The nodes were assigned colors as follows: (blue) authentic HPP enzymes co-localized with other genes involved in the biosynthesis of l-histidine; (green) gene products that possess all the sequence motifs characteristic of the HPP enzymes, but not found co-localized with other l-histidine biosynthetic genes present in these organisms; (red) gene products that possess all the sequence motifs characteristic of an authentic HPP, but the organism lacks a majority of the genes required for l-histidine biosynthesis; (gray) protein sequences that bear significant sequence similarity to authentic HPPs, but lack certain sequence elements critical for HPP activity; (orange) proteins that are not HPP enzymes. Specific HPP enzyme referred to in this paper are indicated by numbers 1–10. These proteins are identified by their locus tags: (1) L37351; (2) MCCL_0344; (3) BBR47_00270; (4) BCE_1533; (5) BcerKBAB4_1335; (6) BSU29620; (7) BH3206; (8) GK2799; (9) SMU_1486c; (10) TTHA0331; (11) LMOh7858_0629. The HPP enzymes characterized in this study are shown as large spheres and enzymes whose crystal structures are available in the PDB are indicated as diamonds. Other crystal structures available for enzymes in this cog are indicated as diamonds and include (A) YcdX from E.coli (pdb: 1m65, 1m68, 1pbo) and (B) N-terminal PHP domain of DNA polymerase X from D.radiodurans (pdb: 2w9m).