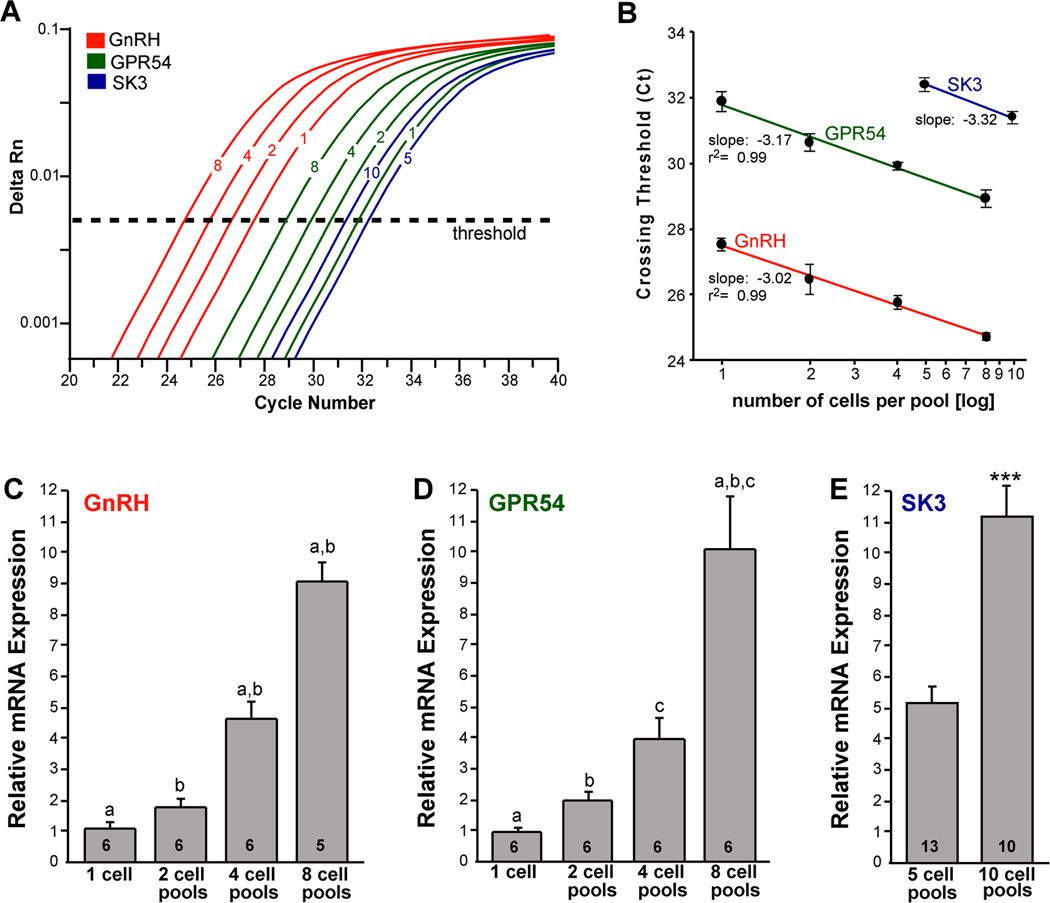
Figure 2.
qPCR amplification for GnRH, GPR54 and SK3 in GnRH neurons. Amplification of cDNA synthesized from differing amounts of RNA in individual GnRH neurons and pools of 2, 4, and 8 cells (GnRH and GPR54) as well as pools of 5 and 10 cells for SK3. cDNA from GnRH single cells and pools was diluted 1:10 for GnRH qPCR measurements. GPR54 and SK3 were quantified in full strength cDNA. A. Cycle number was plotted against normalized fluorescence intensity (delta Rn) to visualize the PCR amplification. Cycle threshold (CT; dashed line) is the point in the amplification at which the sample values were calculated. B. Linear regression analysis plotting crossing threshold (CT) versus the log scale of number of cells collected in each pool (mean ± SEM; n = number of individual cells or pools of cells, which were 5–6 for GnRH and GPR54, and 10–13 for SK3. GnRH, GPR54 and SK3 produced slope values of −3.02, −3.17, and −3.23 respectively, which corresponds to a doubling between each cycle with high linearity (Pearson’s coefficient r2= 0.99). β-actin showed a similar linear relationship between individual cells and cell pools (slope −3.00, r2 = 0.98; data not shown). The amplification efficiency for each primer pair is listed in Table 1. These efficiencies allowed us to use the ΔΔCT method for quantification. C. Bar graphs illustrating the quantitative analysis of relative mRNA expression of GnRH in individual GnRH neurons and in pools of 2, 4 and 8 cells (one-way ANOVA; a-a, b-b, p<0.001; n=6 individual cells and 5–6 pools). D. Bar graphs illustrating the quantitative analysis of relative mRNA expression of GPR54 in individual GnRH neurons and in pools of 2, 4 and 8 cells (one-way ANOVA; a-a, b-b, c-c, p<0.001; n = 6 individual cells and 6 pools). E. Bar graphs illustrating the quantitative analysis of relative mRNA expression of SK3 in GnRH neuronal pools containing 5 and 10 cells (Student’s t-test; ***, p<0.001, n = 10–13 pools).