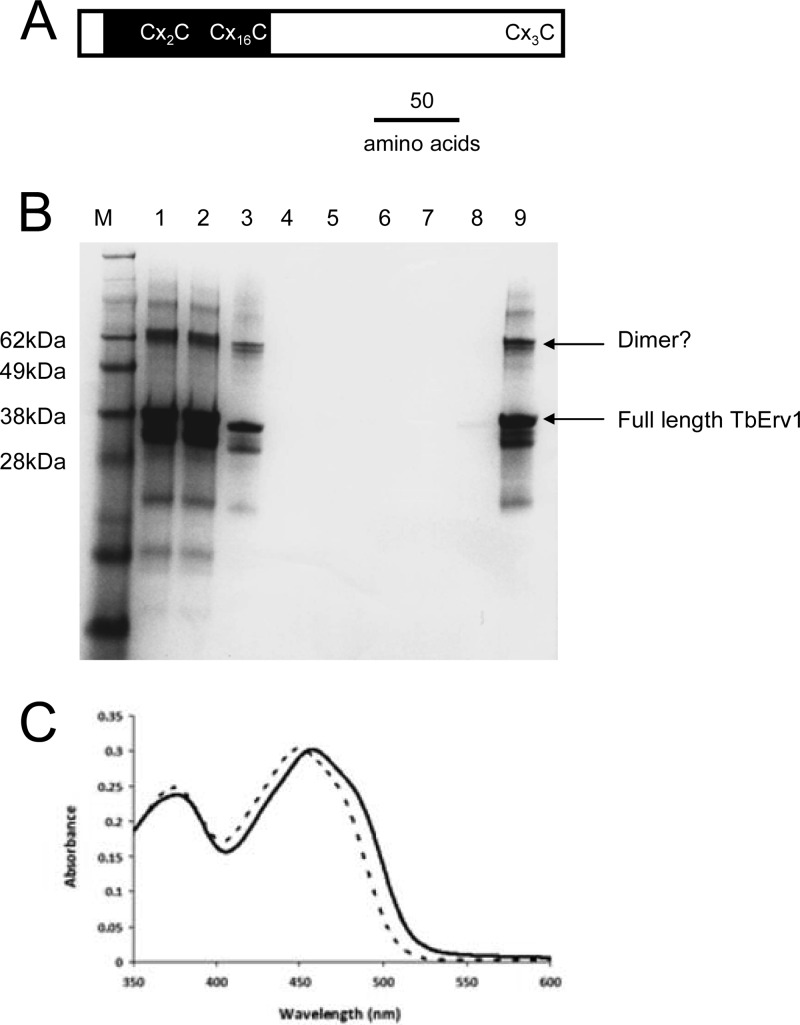
Fig 2.
Isolation of soluble recombinant TbERV1. (A) Schematic of TbERV1 architecture; the relative positions of the conserved core domain (black shading) containing the flavin-binding site and conserved Cys pairs are highlighted. (B) SimplyBlue-stained SDS-PAGE gel documenting in vitro folding of recombinant TbERV1. Lanes: M, molecular mass markers; 1, resuspended high-performance liquid chromatography (HPLC)-purified freeze-dried protein; 2, guanidine–β-mercaptoethanol solubilized protein; 3, solution from lane 2 following incubation with FAD and postdialysis; 4, flowthrough from the Ni-affinity column; 5, 20 mM imidazole–300 mM NaCl–10 mM β-mercaptoethanol wash; 6, 20 mM imidazole–50 mM NaCl–10 mM β-mercaptoethanol wash; 7, 20 mM imidazole–50 mM NaCl wash; 8, 100 mM imidazole–50 mM NaCl wash; 9, 300 mM imidazole–50 mM NaCl elution. Electrospray MS revealed major protein masses at 32,522 ± 6 Da and 29,396 Da; the calculated mass of His6-tagged TbERV1 minus the initial Met is 32,519 Da. Even after the extensive purification and refolding protocol, some low-molecular-mass TbERV1 degradation products remain. (C) Visible absorption spectra of folded TbERV1 and free FAD; dashed line, free FAD; solid line, TbERV1. Absorption maxima of free flavin at ∼375 nm and ∼450 nm shifted to 458 nm for TbERV1, which, together with the shoulder of the 458-nm peak, indicates protein-bound flavin. Flavin occupancy in folded TbERV1 was estimated at 100%, and bleaching of FAD absorption by dithionite treatment confirmed the flavin was redox active.