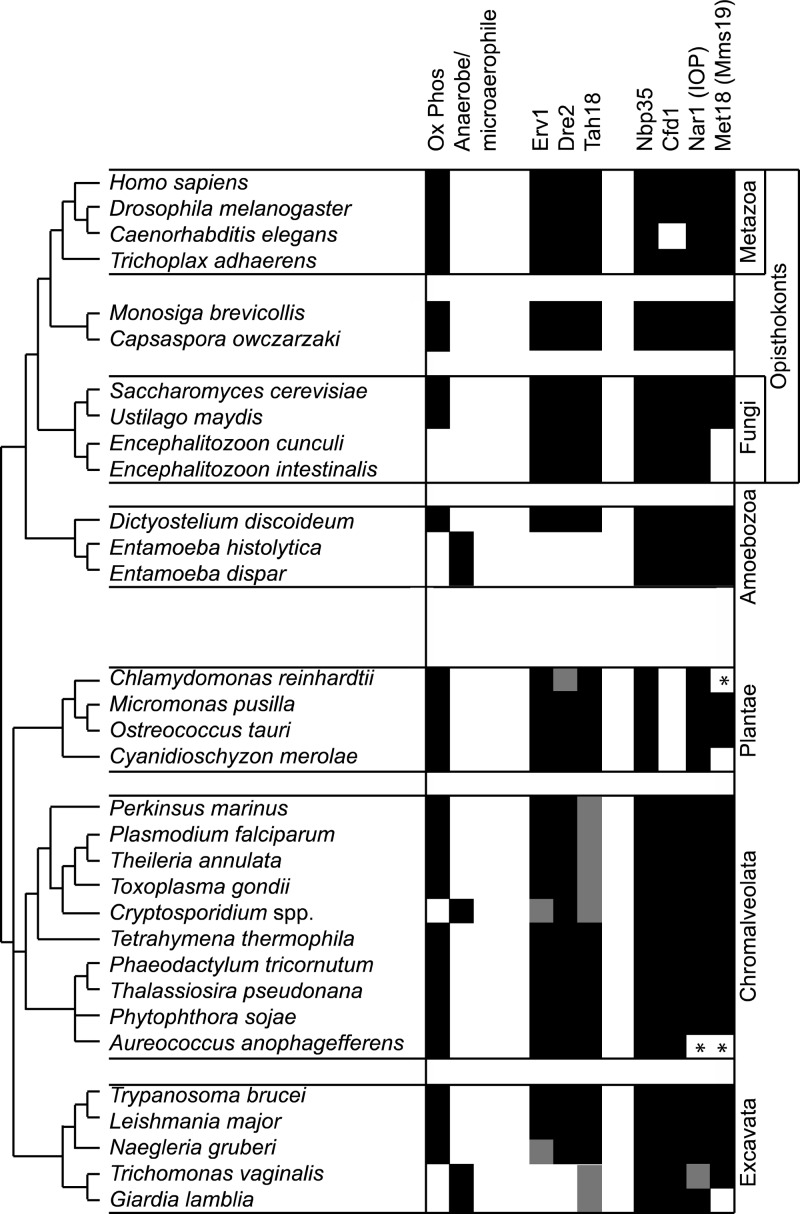
Fig 6.
Phylogenomic distribution in eukaryotes of Erv1 and known components of the CIA pathway. Relationships between different eukaryotic groups represent a consensus of current opinion (11). Conservation of candidate orthologs is denoted by black shading, absence of shading denotes the absence of candidates, and gray shading indicates where there is a clear deviation from a simple binary classification of presence or absence (e.g., atypical domain architecture). Asterisks mark unexpected absences that probably reflect gaps in draft genome coverage. For example, in C. reinhardtii, no Dre2 is predicted from the genome assembly, but Dre2 is present in the closely related Volvox carteri, and a Chlamydomonas expressed sequence tag (EST) with a sequence match to Dre2 is available. For Tah18, homologs lacking conservation to all three domains of the yeast protein are evident in the dinoflagellate Perkinsus marinus, apicomplexan parasites, and Giardia. Curiously, Erv1 can only be found in Cryptosporidium muris, but not C. hominis or C. parvum; a partial hit to Erv1 is evident only from tBLASTn searches in Naegleria gruberi, suggesting problems in prediction of a gene model due to introns. In Trichomonas, canonical-looking Nar1 and Tah18 are not present, but a novel architecture of Nar1 fused to Tah18-related domains is seen.