Fig 1.
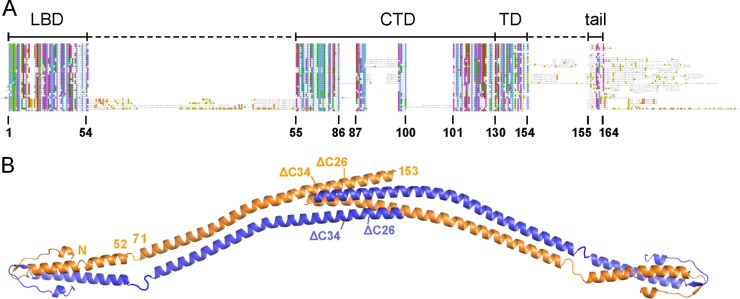
Domain arrangement of B. subtilis DivIVA. (A) Schematic sequence alignment of DivIVA proteins of different phylogenetic origins. Abbreviations above the alignment label the individual protein regions: LBD, lipid binding domain; CTD, C-terminal domain; TD, tetramerization domain; tail, C-terminal tail region. Amino acid numbering is according to the B. subtilis DivIVA sequence. (B) Model of the crystal structure of the full-length B. subtilis DivIVA tetramer which has been assembled from the individual crystal structures of the N- and the C-terminal domains (18). Crystallographic data for the linker between both domains (residues 53 to 70) are not available. Amino acid positions at the beginning and the end of the lipid binding domain as well as the C-terminal domain are indicated for one molecule. Truncation sites of DivIVAΔC26 and DivIVAΔC34 at positions 138 and 130, respectively, are also shown (compare Table 3).
