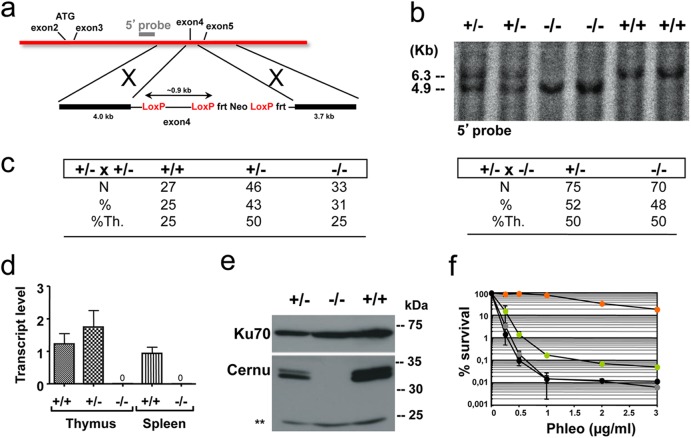
Fig 1.
Generation of Cernu−/− mice. (a) Schematic representation of the KO strategy and location of the external 5′ probe. Exon 4 was flanked by LoxP sites. (b) Southern blot analysis of tail DNA by use of a 5′ probe. The blot shows the 6.3-kb wt band and the 4.9-kb KO allele on a BclI digest. (c) Mendelian distributions of genotypes in offspring of Cernu+/− × Cernu+/− and Cernu+/− × Cernu−/− intercrosses. %Th., percent theoretical. (d) Quantitative RT-PCR evaluation of Cernunnos transcript levels in thymuses and spleens of +/+, +/−, and −/− littermates. (e) Western blot analysis showing the absence of the 35-kDa Cernunnos protein in MEFs. Anti-Ku-70 antibody was used as a loading control. **, nonspecific band. (f) Phleomycin sensitivity of Cernu−/− (green circles), Xrcc4−/− (gray circles), DNA Lig4−/− (black circles), and wt (orange circles) MEFs. The data represent one of two separate experiments, with values representing the means of two independent determinations for each point.