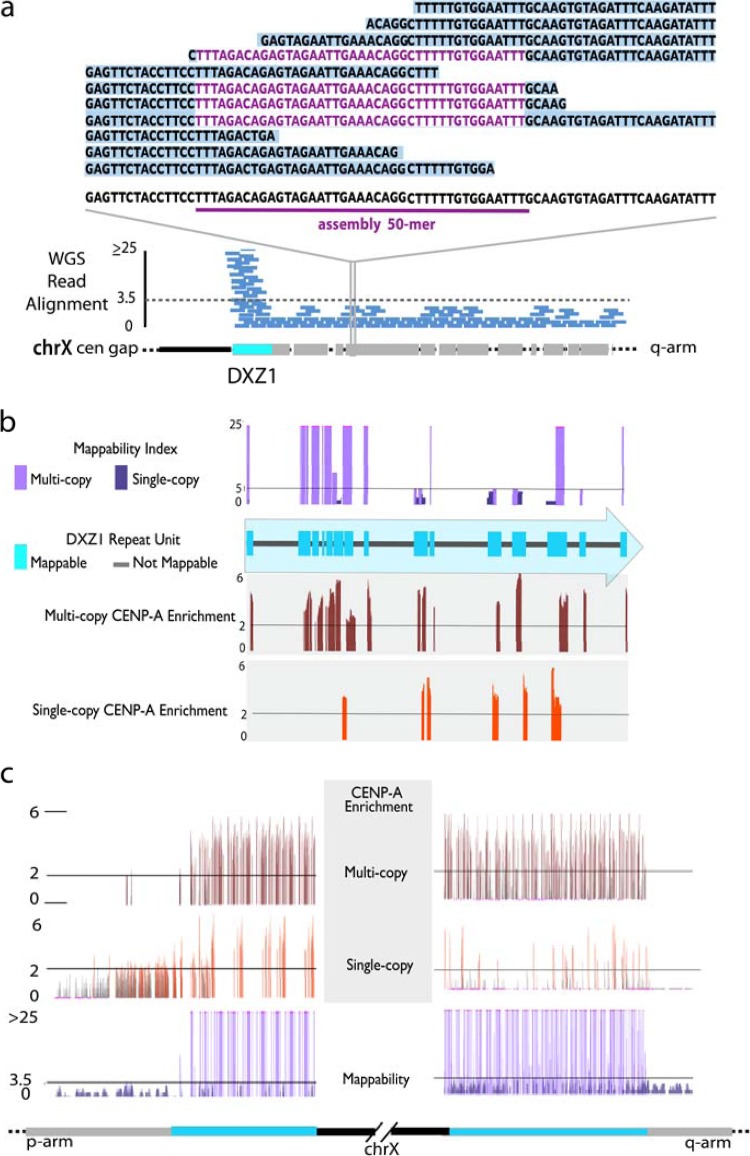
Fig 3.
Characterization of mappable sites of CENP-A enrichment at endogenous human centromeres. (a) HuRef WGS read alignments provide an estimate of read depth, or copy number, as well as provide a database of sequences that are assigned to a particular hg19 annotation. Mappability is illustrated as the comparison of those hg19 annotated 50-mers relative to the assigned HuRef WGS sequence library. This is most clearly demonstrated for those sites suspected to be single copy by read depth estimates, as a given 50-mer is expected to be represented completely by those sequence reads that are assigned to that region. Multicopy sites are defined as mappable if the 50-mer is also identified outside a given location. (b and c) Mapping of CENP-A-enriched 50-mers to these specific mappable sites (shown in teal) can be divided into single-copy and multicopy sites, as defined by read depth. As an example, in DXZ1, we observed both multicopy and single-copy sites within each copy of the higher-order repeat (b) and throughout the assembled portions of the array (c).