Fig 5.
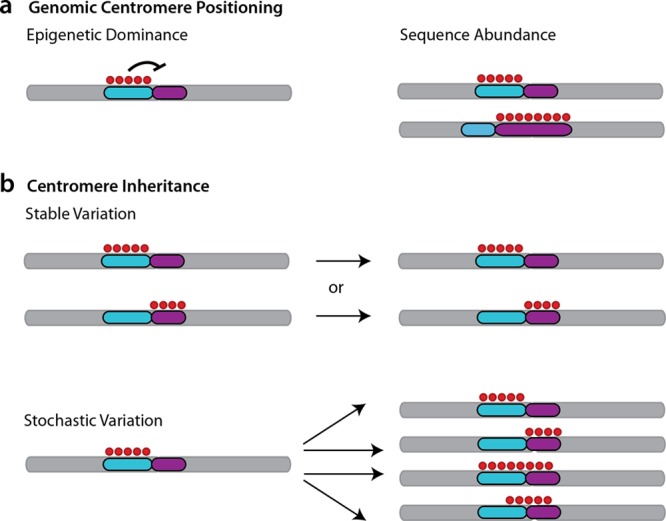
Models for designation of functioning centromere sequences. (a) Genomic features underlying the positioning of functioning centromeres. In an epigenetic model of centromere positioning, the pattern of CENP-A and other marks (red nucleosomes) can act as a positive or negative signal to act upon (and distinguish between) underlying competent genomic sequences (here, purple or teal arrays). Alternately, satellite feature abundance may be a factor contributing to CENP-A recruitment where ancient or more recent polymorphic differences in individual sizes of higher-order arrays influence centromere positioning. (b) Propagation of centromere designation. Under either model of localization shown in panel a, additional models must account for centromere inheritance (mitotic and/or meiotic) and propagation over time. Once loaded, CENP-A association might be stably maintained through mitosis and/or meiosis via nucleosome loading. Conversely or less frequently, these sites might be susceptible to stochastic variation whereby CENP-A enrichment is a signature of a given time point, cell type, or family member but is subject to change and dispersal across all regional competent sequences in other cellular, family, or population contexts.
