Fig 10.
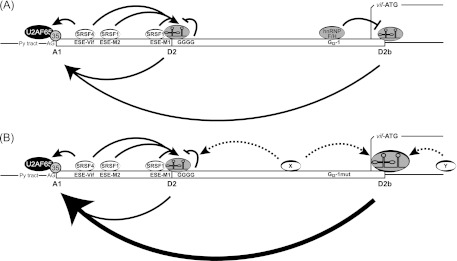
Splicing regulatory model for GI2-1. (A) Splicing at 5′ss D2 and D2b and 3′ss A1 and processing of vif mRNA are modulated by the formation of bridging interactions across exons 2 and 2b. SRSF1-dependent heptameric ESEs M1 and M2 (ESEM1/2) and SRSF4-dependent ESE Vif (ESE-Vif) facilitate the recognition of exon 2. The intronic G runs GGGG and GI2-1 negatively regulate exon 2 and 2b inclusion and levels of vif mRNA. (B) Inactivation of GI2-1 increased the usage of 5′ss D2b and facilitated the recognition of 3′ss A1. The increased usage of the intrinsic weak 5′ss D2b in the absence of GI2-1 could be due to the wide-range effect of SR proteins located upstream (X) and/or downstream (Y) of 5′ss D2b. Py tract, polypyrimidine tract.
