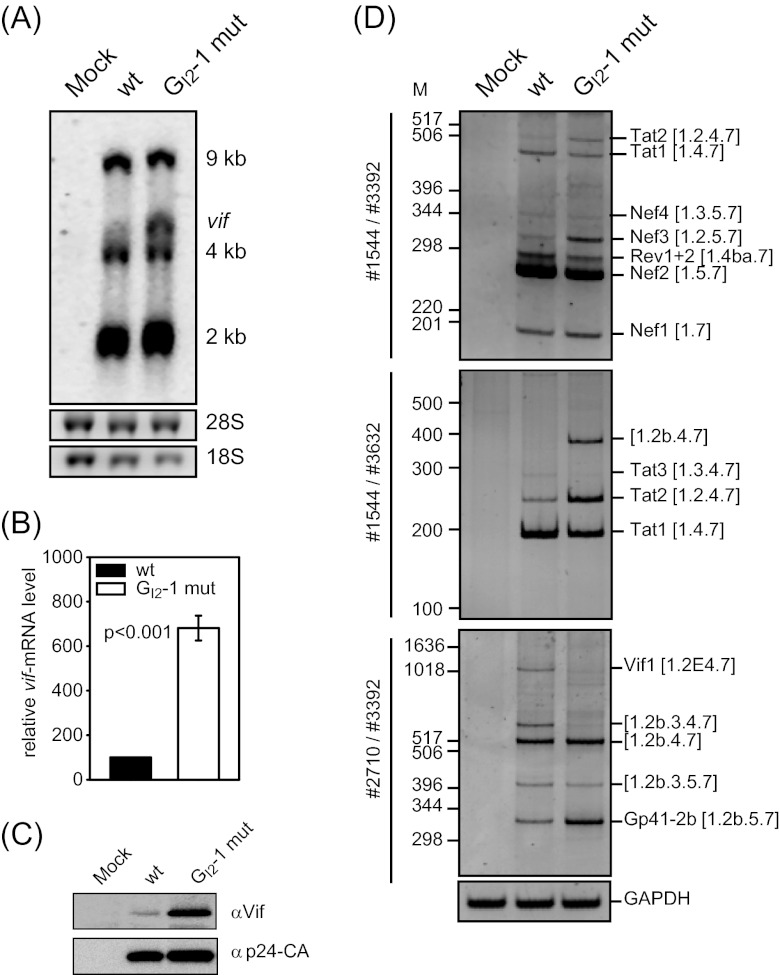
Fig 4.
The silent mutation of GI2-1 increases both vif mRNA and protein levels. (A) Northern blot analysis of total RNA from pNL4-3-transfected HEK 293T cells isolated at 48 h posttransfection. RNA was separated in a 1% RNA agarose gel, capillary blotted, and cross-linked on a positively charged nylon membrane and UV cross-linked. The membrane was treated with a DIG-labeled DNA fragment binding to exon 7. (B) RNA from panel A was subjected to quantitative RT-PCR analysis using an exon junction primer pair specific for vif mRNA (3395/3396) and for exon 7 (3387/3388) for normalization of the viral load. (C) Immunoblot analysis of proteins from transiently transfected HEK 293T cells with proviral plasmids pNL4-3 and pNL4-3 GI2-1 mut. Proteins were separated by SDS-PAGE, blotted, and analyzed with antibodies specific to Vif and p24 as a loading control for the viral load. (D) RT-PCR analysis of total RNA isolated from panel A with primer pairs specific for the 2-kb mRNA class. The primer pairs used are indicated on the left (see Fig. 1). PCR amplicons were separated on a nondenaturing 10% polyacrylamide gel and stained with ethidium bromide. Transcript isoforms are depicted on the right. To compare the total RNA amounts, a separate RT-PCR was performed with the primer pair 3502/3503 to amplify the GAPDH sequence.