Fig 7.
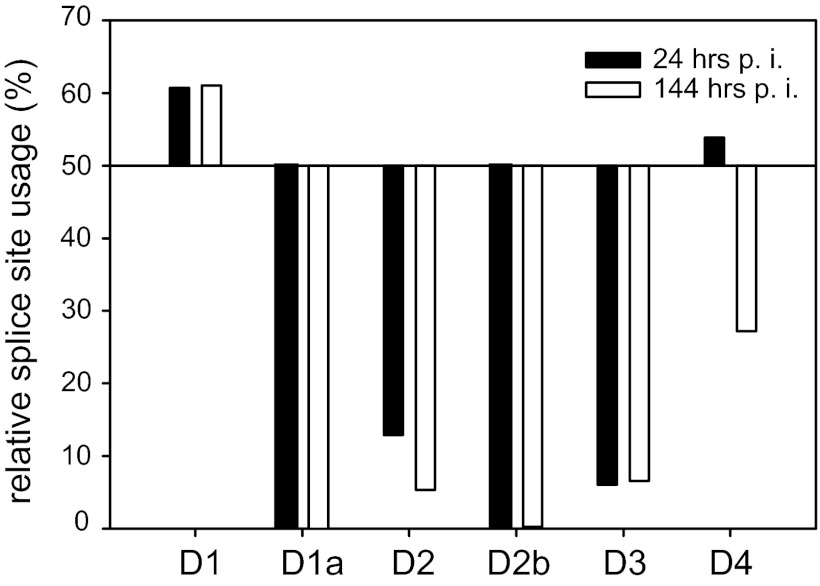
NGS analysis reveals usage of alternative 5′ss D2b. Total RNA of HIV-1 NL4-3-infected Jurkat cells was isolated and subjected to NGS analysis. The resulting 101-nt sequence reads were mapped to the HIV-1 exon junction database, and the relative usage of every splice site was estimated. Read sequences that mapped to more than one exon junction were excluded from quantification. Shown is the relative splice site usage in samples obtained at 24 and 144 hpi.