Fig 5.
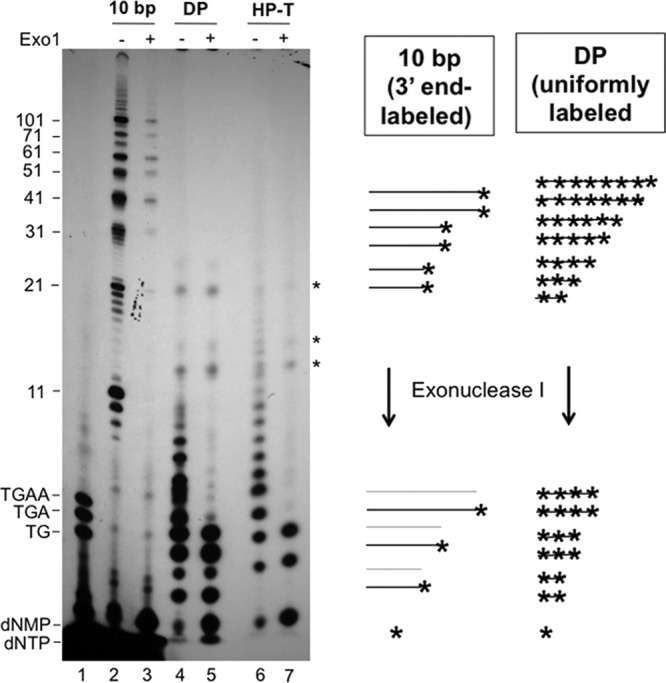
Analysis of protein-primed DNA synthesis products by limited Exo I digestion to differentiate in vitro end labeling of DNA synthesized in vivo versus de novo in vitro initiation and extension (i.e., uniform labeling). The 10-bp marker (Invitrogen) was 3′-end labeled with [α-32P]dATP by Taq polymerase (see Materials and Methods) in a terminal transferase reaction, thus creating 32P-labeled 11-nt, 21-nt, etc., products (lanes 2 and 3). DP was primed in TMnNK buffer in the presence of all four [α-32P]dNTPs, creating uniformly labeled DNA products that were then released by Tdp2 (lanes 4 and 5). HP was primed with [α-32P]TTP, and the HP DNA synthesis products were released by Tdp2 (lanes 6 and 7). All DNA species were then mock treated (−) (even-numbered lanes) or treated with Exo I (+) (odd-numbered lanes, except for lane 1) before resolution by urea-PAGE. The 5′-end-labeled DNA oligonucleotides (dTG, dTGA, and dTGAA) were loaded in lane 1. The positions of the DNA markers are indicated, as are the positions of the dNMPs and dNTPs. *, unknown species resistant to Exo I, apparently bound to protein A/G beads (in both the HP and control DP reactions). These could represent labeled RNAs or peptide fragments. The diagram to the right depicts the expected results from the Exo I digestion of the control, 3′-end-labeled 10-bp DNA markers and the uniformly labeled DP priming products. The asterisks denote the radiolabels at the 3′ ends of or uniformly distributed throughout the DNA molecules. The gray lines denote the DNA molecules that were no longer radiolabeled after removal of the 3′-end nucleotide by Exo I.
