Fig 6.
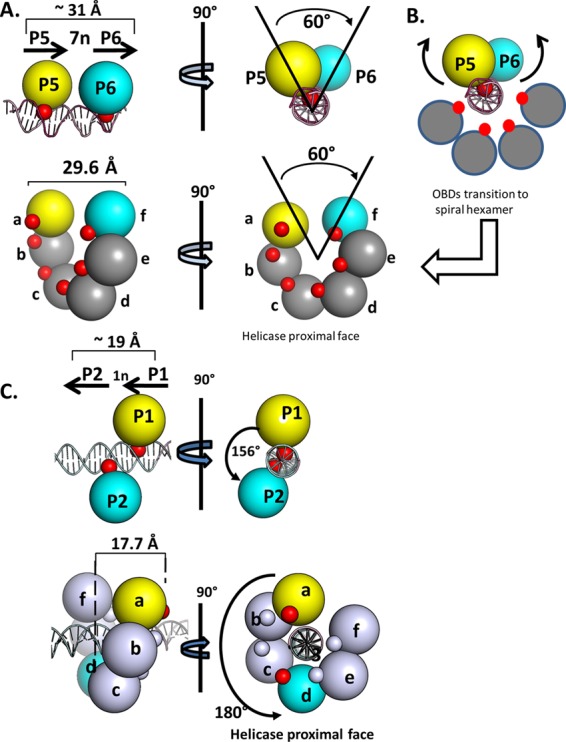
Simplified renderings of the OBD interactions with pairs of pentanucleotides. (A, top) Two views of the OBD interactions with site I. The OBDs are represented by spheres centered at the geometric center of mass. The T-ag OBDs are yellow on P5 and cyan on P6. The smaller red spheres represent the DNA-binding A1 and B2 motifs. (Bottom) Rendering of the left-handed spiral in which the OBDs proximal to the gap are represented by spheres colored as described above. This depiction serves to further illustrate that spiral subunits a and f have the same spatial relationship as the OBDs bound to P5 and P6. As in the top images, the red spheres indicate the relative locations of the A1 and B2 motifs. (B) Modeling studies indicate that the OBDs initially bound to P5 (yellow) and P6 (cyan) undergo a significant transition during spiral formation. This involves rearrangement of the A1 loop from the bound to the free form, subsequent rotation and translation away from the major groove of the dsDNA, and interaction(s) with other OBDs (reviewed in reference 13). (C, top) Two views of the OBD interactions with site II, rotated by ∼90°, used to illustrate the generality of the relationship (24, 26) (RCSB PDB ID 2ITL). P1 and P2 are separated by a 1-bp spacer; therefore, the OBDs bound to P1 (yellow) and P2 (cyan) are ∼156 degrees apart and separated by a translation of ∼19 Å. (As in previous examples, the DNA-binding A1 and B2 motifs are red and magenta, respectively). (Bottom left) Side view of the left-handed OBD spiral hexamer with DNA modeled within the central channel (the 6 subunits are labeled a to f). It is apparent that in this instance, spiral subunits a and d (yellow and cyan, respectively) have a spatial relationship that is analogous to that of the OBD subunits bound to P1 and P2. (Bottom right) Helicase-proximal view of the left-handed spiral structure with duplex DNA modeled in the central channel.
