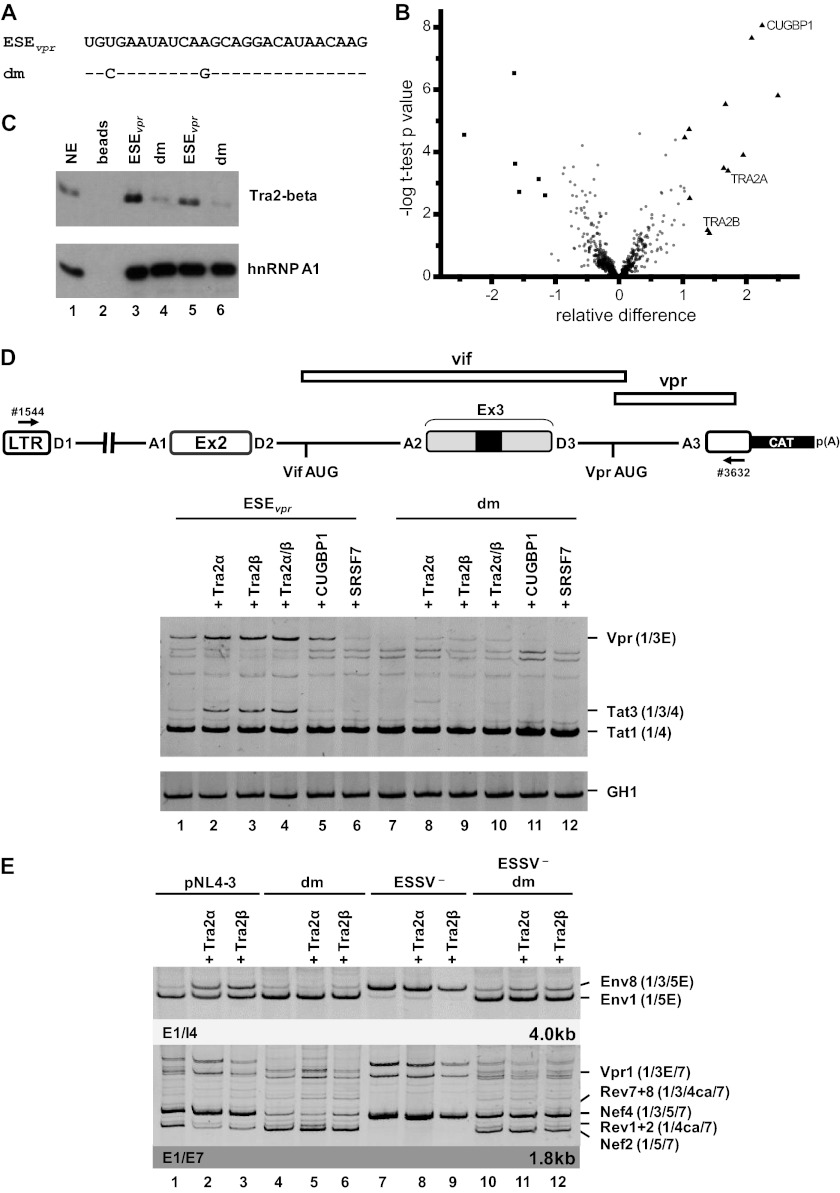
Fig 3.
ESEvpr is bound by the splicing factors Tra2-alpha and Tra2-beta. (A) In vitro-transcribed RNA substrates used for RNA pulldown experiments (dm, double mutation). (B) Volcano plot of RNA binding proteins purified by RNA pulldown with a nonmutated or a mutated ESEvpr sequence with HeLa cell nuclear extract. The precipitated proteins were digested with trypsin and subjected to quantitative mass spectrometry analysis. The x axis of the volcano plot shows the relative difference in protein abundance as calculated by the SAM method, whereas the y axis shows the −log t-test P value of the groupwise comparison of protein abundances. Besides the majority of probably unspecifically binding proteins (circles), some proteins preferably bound to the wild-type ESEvpr sequence (triangles) or the mutated ESEvpr variant (squares). The proteinsTra2-alpha and Tra2-beta were selected for validation experiments. (C) Immunoblot analysis with an antibody specific for Tra2-beta and hnRNPA1 confirmed significantly smaller amounts of Tra2-beta for the double mutant. (D) HeLa cells (2.5 × 105) were transiently cotransfected with 1 μg of each of the HIV-1-based LTR ex2 ex3 splicing reporters, 0.2 μg of SVctat (47); 1 μg of pXGH5 (GH1) as a transfection control, and 1 μg of pcDNA3.1(+), an expression plasmid for Tra2-alpha, Tra2-beta, CUGBP1, and SRSF7. At 30 h posttransfection, total RNA was isolated and subjected to semiquantitative RT-PCR analyses with primers 1544 and 3632. For measurement of equal transfection efficiencies, a separate PCR was carried out with a primer pair (1224/1225) specific for human growth hormone 1 (GH1). (E) RT-PCR analyses of intronless (2-kb) and intron-containing (4-kb) viral mRNA species following the transient transfection of HEK 293T cells with 1 μg of the respective proviral construct and 1 μg of pcDNA3.1(+), an expression plasmid for either Tra2-alpha or Tra2-beta. E, extended exon; dm, double mutation.