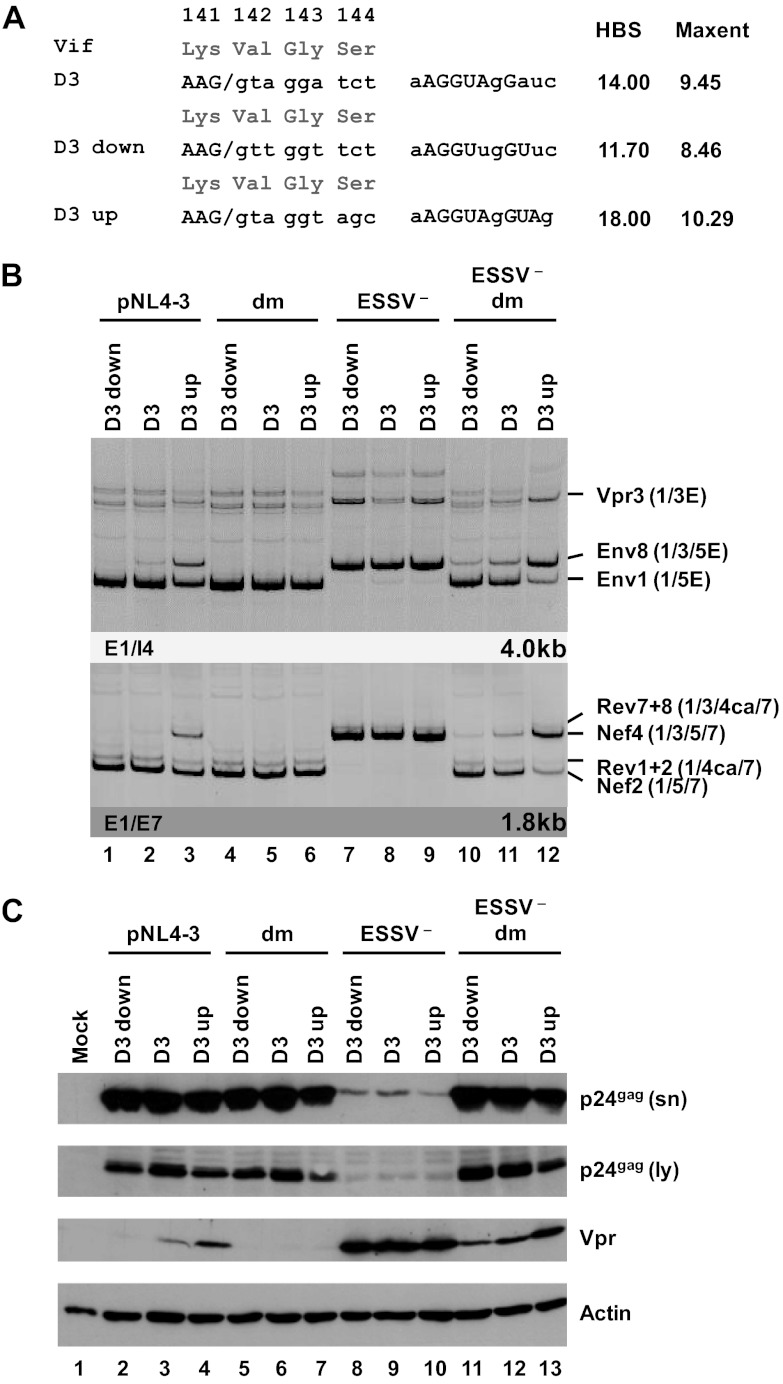
Fig 5.
5′ss D3 up and down mutations modulate HIV-1 exon 3 splicing and vpr mRNA formation. (A) Silent mutations predicted to decrease or increase the complementarity to the 5′ end of the endogenous U1 snRNA were introduced into viral 5′ss D3. Exonic nucleotides are denoted in uppercase letters, and intronic nucleotides are denoted in lowercase letters. Complementarity and predicted intrinsic strength by HBond score (HBS) and MaxEnt score algorithms are both shown next to the 5′ss sequence. Nucleotides complementary to the U1 snRNA are in capital letters, while mismatches to the U1 snRNA are in lowercase letters. (B) HEK 293T cells (2.5 × 105) were transiently transfected with 1 μg of each of the different infectious clones. RNA was isolated from the cells, DNase I digested, and reverse transcribed. The resultant cDNA served as the DNA template in semiquantitative PCRs using primer pairs E1/I4 and E1/E7 to specifically detect viral 4.0- and 1.8-kb viral mRNAs, respectively. Proviral mutants are shown above the panels. The main HIV-1 mRNA species are indicated at the right. (C) Protein lysates and viral supernatants were collected from HEK 293T cells transfected with 1 μg of pNL4-3 or mutant derivatives. Samples were loaded on 12% SDS-polyacrylamide gels and, after separation, transferred to nitrocellulose membranes. Viral proteins and α-actin (as a loading control) were determined by probing with specific primary antibodies. For detection, appropriate HRP-conjugated antibodies and ECL detection reagent were applied. HBS, HBond score; MaxEnt, MaxEnt score; dm, double mutation; E, extended exon; sn, supernatant; ly, lysate.