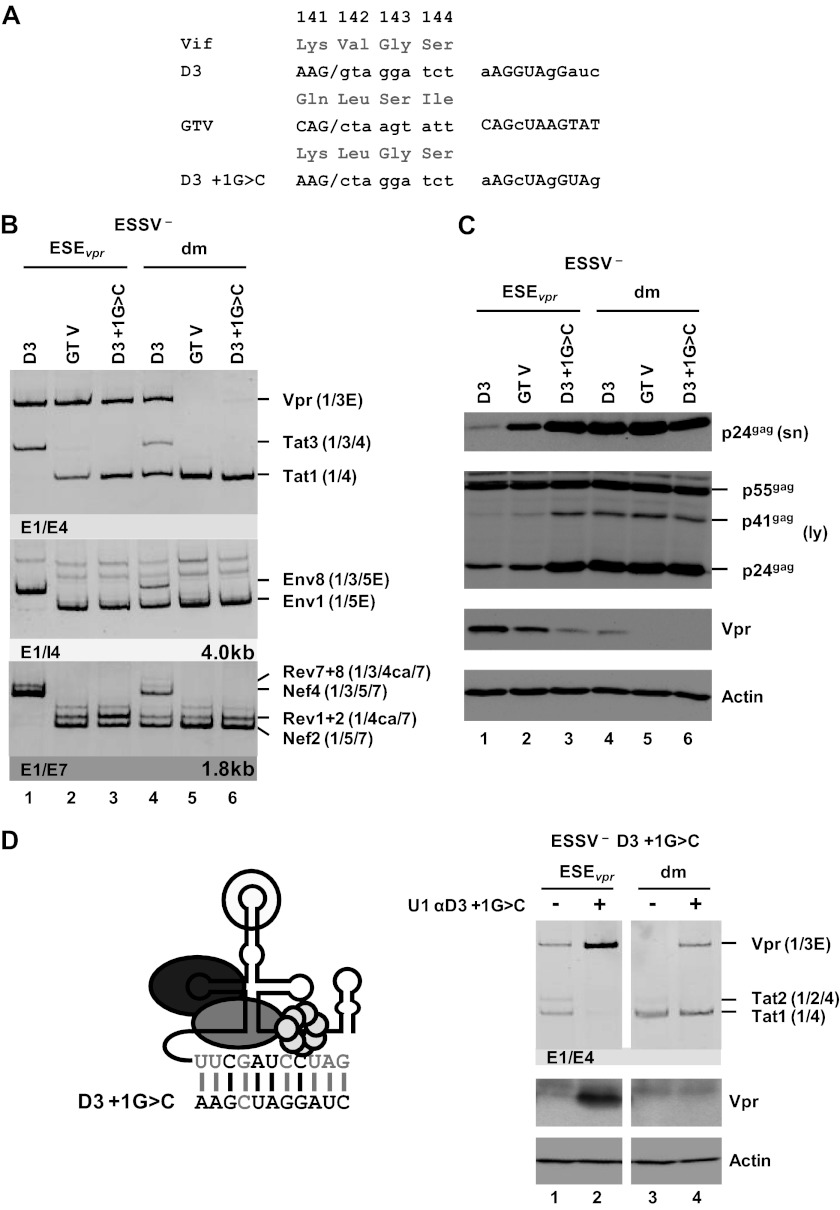
Fig 6.
U1 snRNP binding to a splicing-incompetent 5′ss enhances vpr mRNA expression. (A) 5′ss D3 was replaced with a splicing-incompetent sequence that perfectly matches the free 5′ end of the cellular U1 snRNA except for position +1 (GTV). As a control, 5′ss D3 was disabled for splicing by a G-to-C mutation at position +1, decreasing its complementarity to the U1 snRNA (D3+1G>C). Complementarity patterns are shown next to the 5′ss sequences. Matches to the U1 snRNA are indicated by uppercase letters, and residues not complementary are in lowercase letters. (B) HEK 293T cells (2.5 × 105) were transiently transfected with 1 μg of each of the proviral constructs and analyzed by semiquantitative RT-PCR. RT-PCR products were resolved by PAGE, followed by ethidium bromide staining. Mutants are depicted at the top. Main viral mRNAs are indicated on the right. (C) Cellular lysates and viral supernatants were obtained from transfected HEK 293T cells and loaded onto 12% SDS-polyacrylamide gels. After transfer to nitrocellulose membranes, viral proteins were determined with specific antibodies for p24gag and Vpr. To ensure the loading of equal protein amounts, the membrane was also probed with an antibody to cellular α-actin. (D) Schematic drawing of a 5′-end-modified U1 snRNA perfectly matching the 5′ss D3+1G>C sequence (left). Mutated nucleotides are indicated by gray capital letters. Additional base pairing interactions between 5′ss D3 and the optimized 5′ end of the U1 snRNA are indicated by vertical gray lines. HEK 293T cells (2.5 × 105) were transiently transfected with 1 μg of both proviral pNL4-3 DNA and U1 snRNA expression plasmid. Total RNA and cellular lysates were isolated and subjected to RT-PCR or Western blot analysis (right). E, extended exon; dm, double mutation; sn, supernatant; ly, lysate.