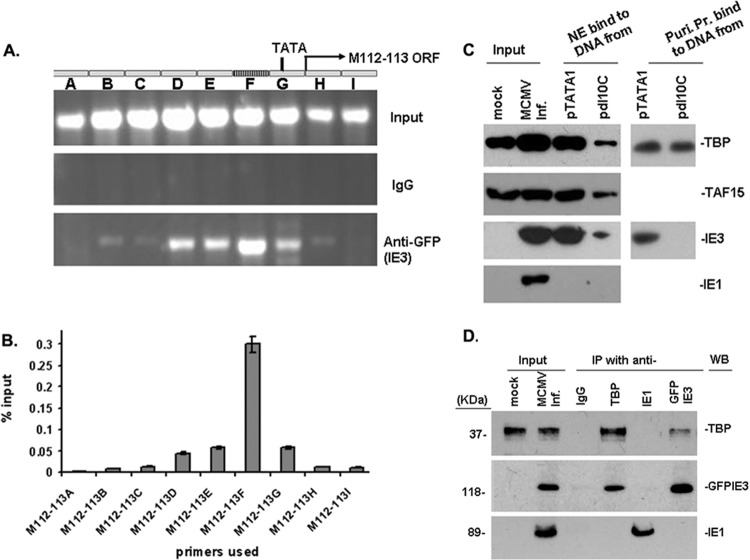
Fig 3.
Ten-bp DNA motif mediates the interaction of IE3 with TBP and stabilizes TFIID complexes. (A and B) DNA ChIP assay to examine IE3-DNA interaction. (A) M112-113 gene structure is shown at the top; input DNA was amplified to show the total DNA in the samples, and IgG-incubated samples were used as a negative control. In the bottom panel, ChIP DNA using anti-GFP shows that IE3-bound DNA is the strongest in the region (F) where the 10-bp motif resides. (B) Real-time PCR was performed to show the percentage anti-GFP ChIP DNA compared to the amount of the input. Results are consistent with those of regular PCR shown in A. (C) DNA affinity assay to ascertain direct interaction of IE3 with the M112-113 promoter with or without the 10-bp DNA. (Left side) DNA affinity assay with the whole nuclear extract (NE). Mock indicates the NE prepared from uninfected NIH 3T3 cells. Input was NE made from MCMV-infected (MCMV Inf.) NIH 3T3 cells. (Right side) DNA affinity assay with purified proteins (Puri. Pr.) of TBP (upper) and GFPIE3 (lower). (D) Coimmunoprecipitation assay to determine the interaction of TBP with IE3. Nuclear extracts were prepared from NIH 3T3 cells infected with MCMVE5gfp at a multiplicity of infection of 1 and incubated with antibodies as indicated at the top of the panel. The pulled-down proteins were detected by Western blotting using antibodies shown on the right side. For the detection of protein in the immunoprecipitation, we used secondary antibodies from TrueBlot ULTRA (18-8817 for mouse or 18-8816 for rabbit; eBioscience) to avoid the heavy chain.