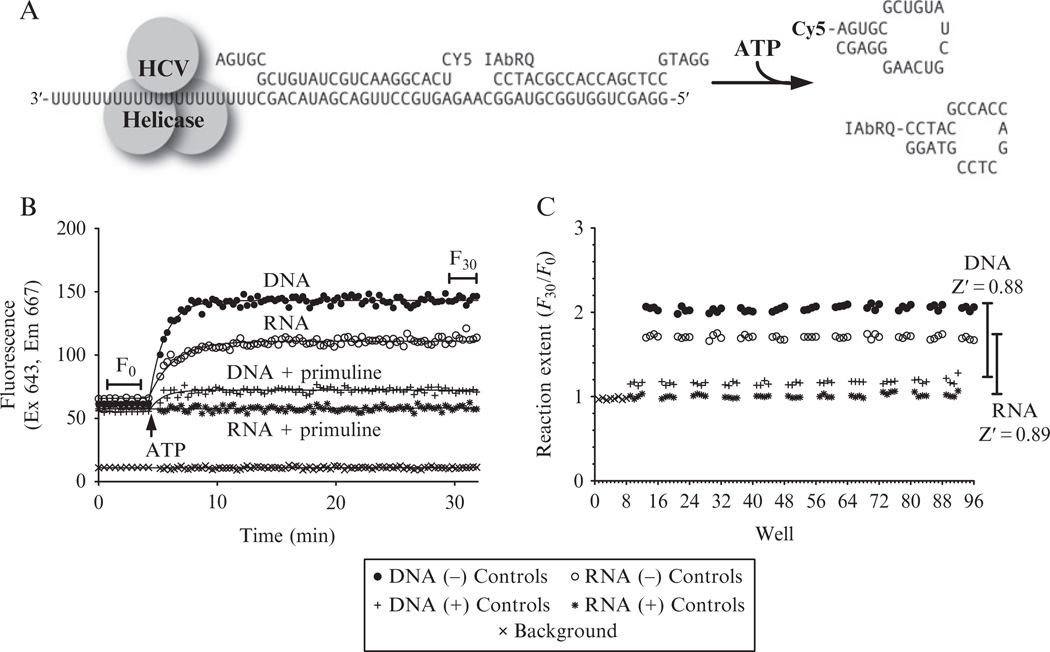
Figure 21.4.
An RNA-based HCV helicase assay suitable for HTS. (A) Design of the split beacon helicase assay (SBHA). In the RNA substrate, the bottom strand and the Cy5-labeled strand are RNA, while the IAbRQ-labeled strand is composed of DNA. In the DNA-labeled substrate, all three strands are composed of DNA, with Ts replacing Us in the bottom strand and the Cy5-labeled strand. (B) Time courses for sample negative control reactions using the DNA (closed circles) and the RNA (open circles) substrate, positive control reactions containing DNA plus 50 µM primuline (+), and the RNA substrate with 50 µM primuline (*). Lines show fit to the equations described in the text. The fluorescence observed in a well without substrate (×) is shown for comparison. (C) Reaction extent for two 96-well plates, one of which contained an RNA substrate and one a DNA substrate.