Figure 8.
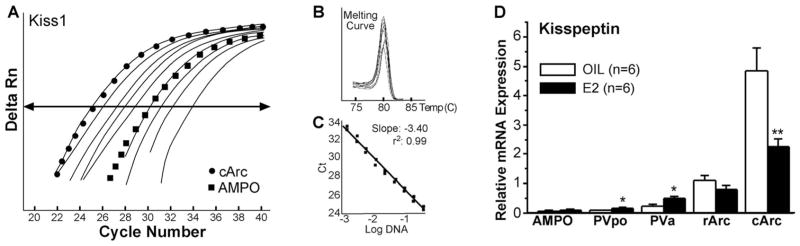
Kiss1 mRNA distribution by real-time PCR. Quantitative real-time PCR measurements of Kiss1 mRNA in microdissected hypothalamic nuclei in oil- and E2-treated female guinea pigs. A standard curve was prepared with basal hypothalamic (BH) cDNA serial dilutions from 1:50 to 1:12,800 (A). Cycle number was plotted against the normalized fluorescence intensity (Delta RN) to visualize the PCR amplification. The cycle threshold (Ct; arrow) is the point in the amplification at which the sample values were calculated. The BH cDNA serial dilutions and two representative samples (cArc and AMPO) are illustrated in A. The superimposed dissociation (melting) curves depict a single Kiss1 product (B). The standard curve regression line and slope from serial dilution data are illustrated (C). The amplification efficiency calculated from the slope was 96% for Kiss1. GAPDH was used as a control, and its amplification efficiency was 98% (data not shown). The expression values were calculated using the ΔΔCt method where the calibrator was the ΔCt of rArc oil-treated samples (D). Number of animals (n) = 6 in each group; *P < 0.05, **P < 0.01, two-tailed Student’s t-test.
