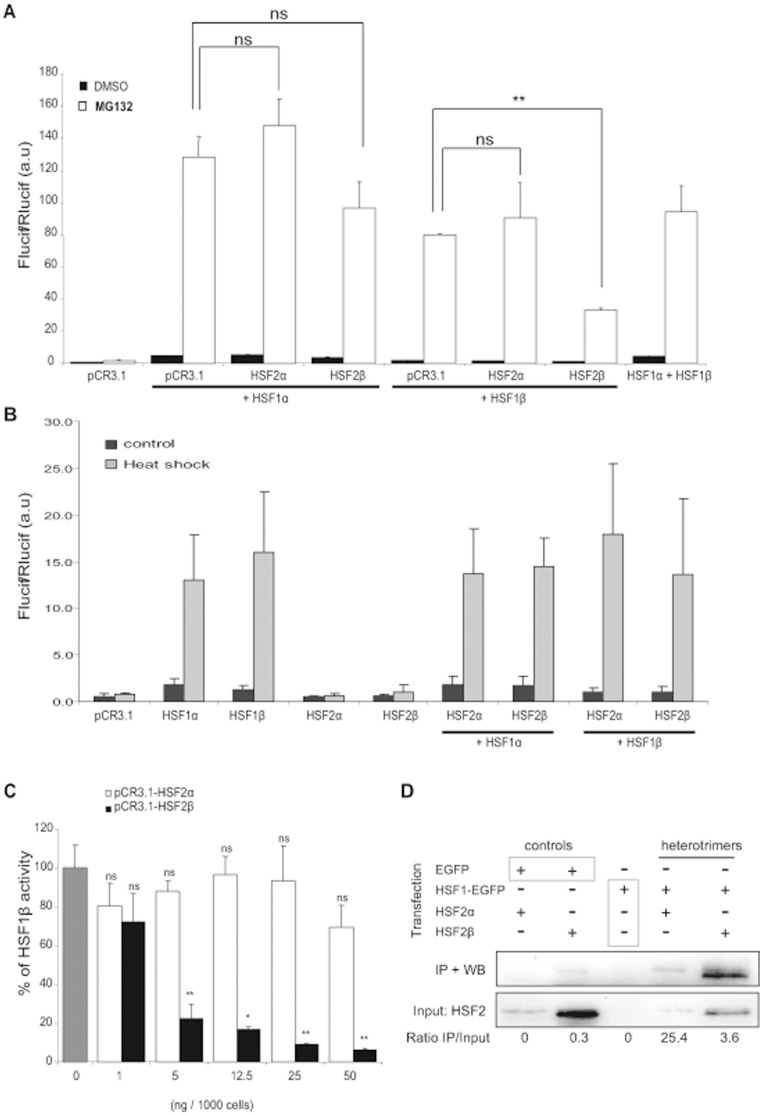
Figure 2. HSF2β interacts with HSF1β and inhibits its transcriptional activity.
(A) Hsf1.2−/− iMEFs were co-transfected with 12.5 ng of pCR3.1-HSF1α or pCR3.1-HSF1β in combination with 12.5 ng of pCR3.1-HSF2α, or pCR3.1-HSF2β. Transcriptional activity was followed with pHSEx2-TATA-Luc, and pTK-Rluc was used as control for transfection efficiency. Cells were treated with MG132 at 2.5 µM (white), or with DMSO as control (black). Results correspond to the ratio between firefly luciferase (FLucif) and renilla luciferase (RLucif) activities and are a representative experiment made with three independent replicates (Student’s t test: HSF1α or β alone compared to others conditions. **p<0.01, ns: no significant). (B) Cells were transfected with the indicated expression vectors and the reporter genes, as described in (A). Cells were submitted to heat shock at 45°C for 20 min and put to recovery at 37°C for 6 h (grey). Untreated cells served as control (black). Results are the mean +/− SD of 9 to 12 independent transfections. (C) Hsf1.2−/− iMEFs were co-transfected with pCR3.1-HSF1β (gray) and with an increase quantity of pCR3.1-HSF2α (white), or pCR3.1-HSF2β (black). Then, cells were treated with MG132 at 2.5 µM for 8 h. Results are expressed in percentage of HSF1β activity and are the mean of three independent experiments +/− SD (Student’s t test: HSF1β alone compare to others conditions. *p<0.05, **p<0.01, ns: no significant). (D) Representative co-immunoprecipitation experiments. Hsf1.2−/− iMEFs were co-transfected with pEGFP or pHSF1β-EGFP, in combination with pCR3.1-HSF2α or pCR3.1-HSF2β. Cells were treated with MG132 at 1 µM for 8 h and nuclear protein extracts were submitted to EGFP immunoprecipitation, followed by immunoblotting for HSF2. The value of the IP/Input ratio is indicated below the panel.