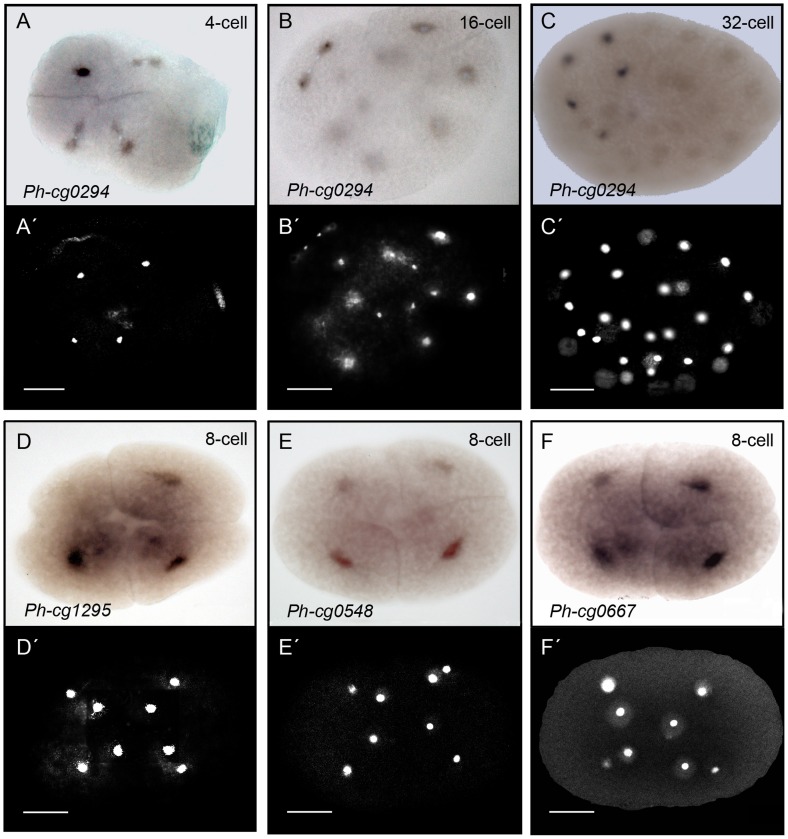
Figure 4. Whole mount in situ hybridization analysis of asymmetrically distributed RNAs at the 8-cell stage of Parhyale.
Shown here four out of the six RNAs that generated signal. The RNA is detected in the small perinuclear cytoplasm at different signal intensities. (A-C) Bright field views and (A′–C′) corresponding DAPI stained images of three different embryos stained for Ph-cg0294 (microarray target: Contig0106) and DNA. The cytoplasmic in situ signal (dark) is concentrated in the precursor of the Mav cell at the 4-cells stage shown in (A) (Mav is the progenitor of anterior visceral mesoderm, 2nd quadrant of image, see Figure 2B). The nucleus of the cells is in the centre (A′), surrounded by a small region of cytoplasm (in situ staining only in cytoplasm). The remaining volume of the blastomeres is yolk-rich. (B, B′) An embryo entering the 16-cell stage and the Mav cell finishing the process of division. The in situ signal does not colocalize with the nuclei of the daughter cells of Mav, but is rather cytoplasmic. Weak signal is also visible in the daughter cells of El (progenitor of left ectoderm, 1st quadrant). (C, C′) A 32-cell stage embryo with strong cytoplasmic staining in four cells (the lineage of Mav) and weaker signal in a fifth cell. (D, D′) An 8-cell stage embryo showing the expression pattern of Ph-cg1295 (microarray target: zaFBplate1_E16_060.ab1). A strong signal was detected in the progenitors of ectoderm and weaker signal was visible in two of the micromeres. (E, E′) The WISH analysis for cg0548 (microarray target: zamn02_K22_085.ab1) shows weak staining in the macromeres but not the micromeres of the 8-cell stage. (F, F′) WISH analysis on 8-cell stage embryos for cg0667 (microarray target: zamn01_E08_028.ab1) showed stronger signal in the ectoderm macromeres, weaker signal in the mesoderm macromere and no signal in the micromere progenitors. Scale bar corresponds to 100 µm.