Figure 1. Evolutionary model and assumptions behind our forward genomics approach.
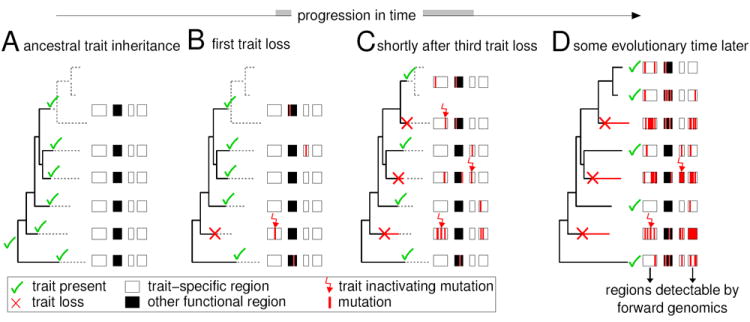
(A) An ancestral trait is passed to descendant species, along with the genomic regions required for this trait, which evolve under purifying selection.
(B) One lineage loses the ancestral trait due to an inactivating mutation in a trait-required region.
(C) Following trait loss, all trait-specific (non-pleiotropic) regions switch to evolve neutrally and begin to accumulate random mutations in the first trait-loss lineage. Meanwhile, two additional independent lineages lose this trait, due to independent mutations occurring either in the same or in other trait-required regions.
(D) All trait-specific regions continue to erode independently in the three different trait-loss lineages, while their counterparts in the trait-preserving species are conserved due to purifying selection. This characteristic evolutionary signature can be detected using forward genomics, revealing functional components of this (monogenic or polygenic) trait.
