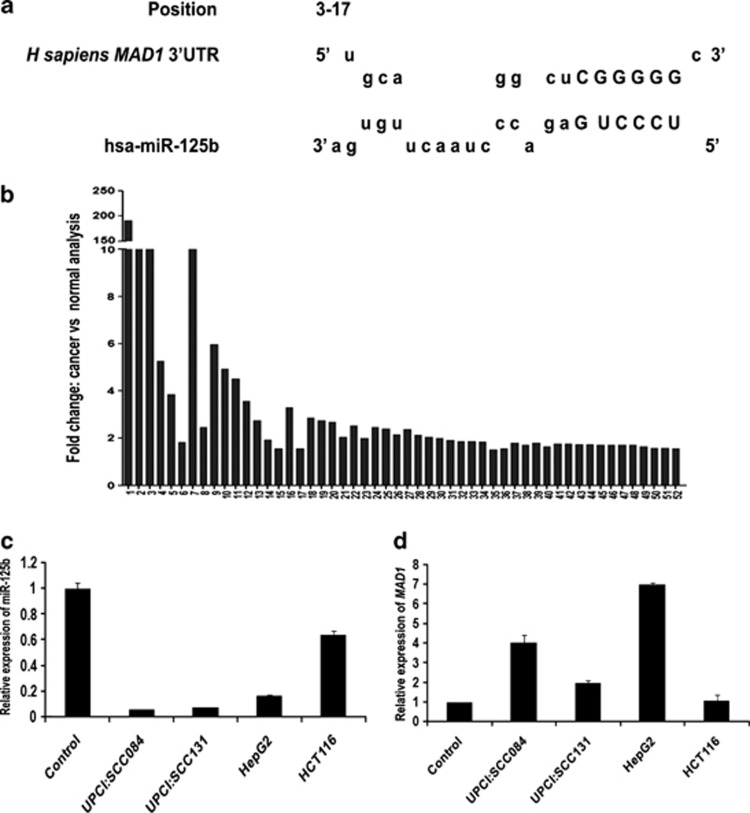
Figure 1.
MAD1 is a putative target of miR-125b. (a) The nucleotide position 3–17 of MAD1-3′UTR is the recognition site for miR-125b as predicted by RNAhybrid. Letters in upper case denote the seed region. (b) Mad1 expression is upregulated in different cancers. Expression data of Mad1 in 52 different cancers from Oncomine database were analysed. Cancer versus normal data sets of Mad1 overexpression with fold change ≥1.5 and P-value≤0.05 were selected. Bars represent fold overexpression of Mad1 in these cancers. (c) Endogenous miR-125b expression in different cancer cell lines as compared with control oral epithelial cells. Total RNA was isolated from oral swab (control), UPCI:SCC084, UPCI:SCC131, HepG2 and HCT116 cells and reverse transcribed using miR-125b and miR-17-5p specific stem-loop primers. cDNAs were subjected to RT-PCR. Relative expression values were normalized to those of miR-17-5p. (d) Corresponding endogenous Mad1 levels in the above cell lines as compared with control oral epithelial cells. Total RNA was reverse transcribed and RT-PCR was done using MAD1-specific primers. Relative expression values were normalized to those of GAPDH. For both (c) and (d), data represent three independent experiments (N=3) and are shown as average±S.E. N=number of times the experiment was performed