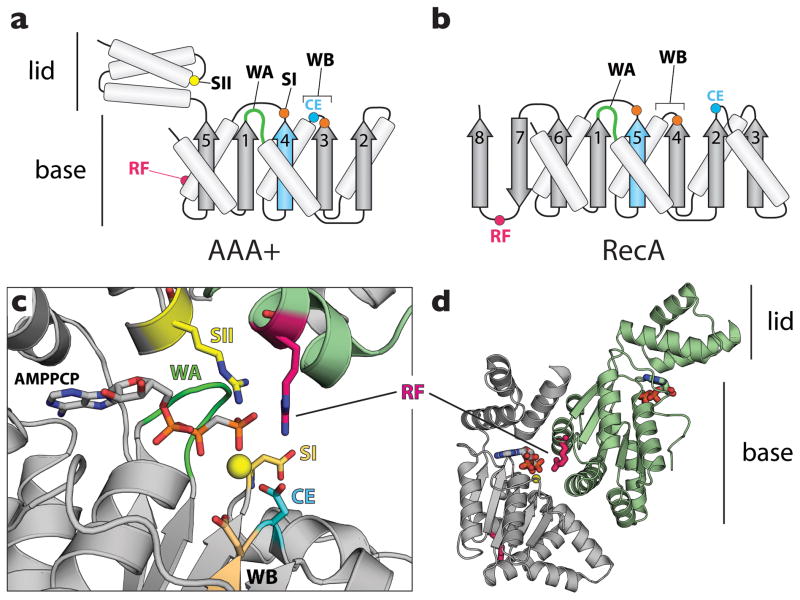
Figure 1.
ASCE ATPase architecture. AAA+ enzymes share a common structural core with RecA-type ATPases, together forming the Additional Strand Catalytic glutamate (E) (ASCE) supergroup of P-loop NTPases [79]. (a) Topology diagram for a AAA+ ATPase. AAA+ proteins frequently possess a nucleotide-binding pocket formed by two subdomains, a base αβ α-nucleotide-binding fold and a C-terminal α-helical lid domain. (b) Topology diagram for a RecA ATPase. In panels a and b, the additional strand and catalytic glutamate are highlighted in cyan. (c) Signature sequence motifs of AAA+ proteins. WA – Walker A (dark green), a glycine-rich element that forms a distinctive loop for cradling the phosphates of ATP. WB – Walker B (cyan and orange), a motif that contains acidic residues that interact with an associated magnesium ion (yellow sphere). SI/SII – sensor I (orange) and sensor II (yellow) elements, which reside near and can interact with the γ-phosphate of ATP. (d) Assembly patterns in AAA+ ATPase oligomers. A typical AAA+ dimer is shown (DnaA in this instance; PDB ID: 3R8F [9]). A conserved arginine residue – the arginine finger (RF, magenta) – extends from one AAA+ subunit (light green) to contact the γ-phosphate of ATP bound to a neighboring protomer (gray).