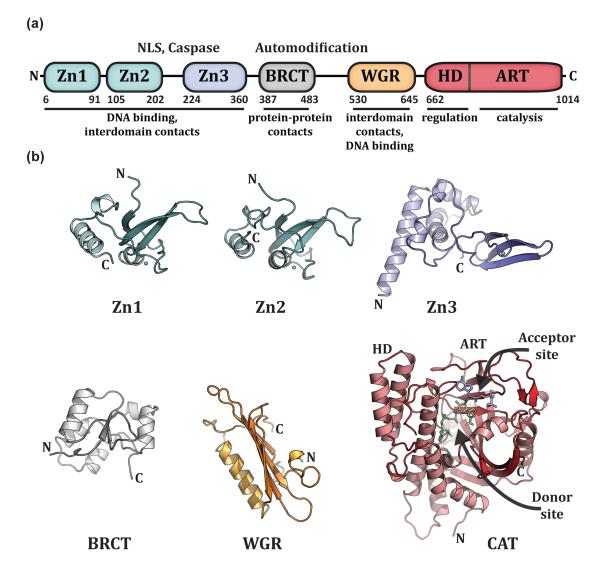
Figure 1. PARP-1 has a highly modular, “beads-on-a-string” architecture.
(a). Schematic representation of human PARP-1 domains. A bipartite nuclear localization signal (NLS) and a caspase 3 cleavage site (Caspase) are located between the Zn2 and Zn3 domains. A BRCA C-terminus (BRCT) fold is located within the region of PARP-1 that is primarily targeted for automodification. The catatlyic domain is composed of an alpha-helical subdomain (HD) and an ADP-ribosyl transferase subdomain (ART). (b). Crystal and/or NMR structures have been determined for each of the PARP-1 domains in the absence of DNA. Shown are the NMR structures of the homologous Zn1 and Zn2 domains (PDB code 2dmj and 2cs2, respectively [64], see also [8]), the NMR structure of the Zn3 domain (PDB code 2jvn, see also reference [9] for crystal structure), the NMR structure of the BRCT fold (PDB code 2cok [64], see also reference [11]), the NMR structure of the WGR domain (PDB code 2cr9 [64]), and the crystal structure of the catalytic domain (PDB code 1a26 [12]). Catalytic subdomains are labeled (HD and ART). The catalytic domain structure was determined in the presence of an NAD+ analogue that has defined the “acceptor site” for poly(ADP-ribose) formation. NAD+ is modeled in the “donor site” based on the related structure of diphthetia toxin [19]. All structure depictions were made using PYMOL (www.pymol.org).