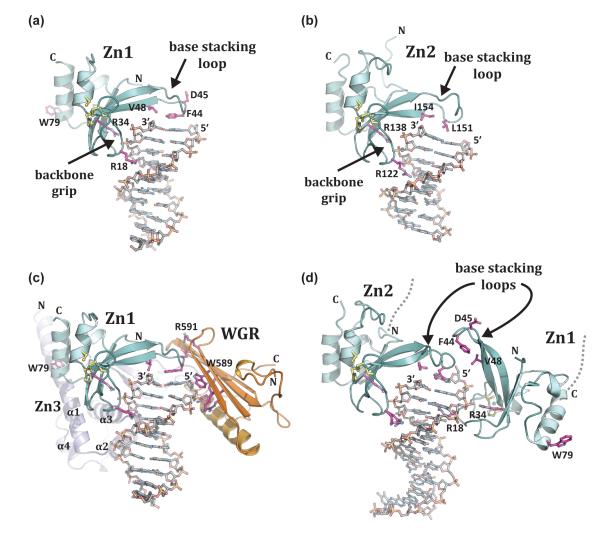
Figure 2. PARP-1 recognizes exposed nucleotide bases as a signature of DNA damage.
(a). Crystal structure of human Zn1 domain in complex with a DNA double strand break (PDB code 3od8 [13]). Together with panel b, these structures illustrate the features of damage DNA that are recognized by PARP-1 zinc finger domains: a continuous phosphate backbone engaged by conserved Arg residues on the “backbone grip”, and exposed nucleotide bases engaged by hydrophobic residues on the “base stacking loop”. D45 and W79 are key Zn1 residues involved in contacts with other PARP-1 domains. (b). Crystal structure of human Zn2 in complex with a DNA double strand break (PDB code 3odc [13]). (c). The Zn1, Zn3, and WGR domains collectively assemble on a DNA double strand break, with each domain forming specific protein-DNA, and protein-protein contacts with adjacent domains [41] (see also Figure 3a). Zn1 has the same orientation as in panel a. Residues critical for PARP-1 DNA-dependent activity are labeled: WGR residues W589 and R591, and Zn1 residue W79. (d). Crystal structure of the tandem Zn1–Zn2 domains of PARP-1 bound to a DNA double strand break with a single 5’ nucleotide overhang [40]. Zn2 forms DNA contacts similar to that seen for the isolated domain bound to DNA in panel b (the same conserved residues are shown). Zn1 binds to DNA with opposite polarity to that seen in panel a, with Arg18 positioned in the major groove rather than the minor groove, and the “base stacking loop” forming protein-protein contacts rather than protein-DNA contacts.