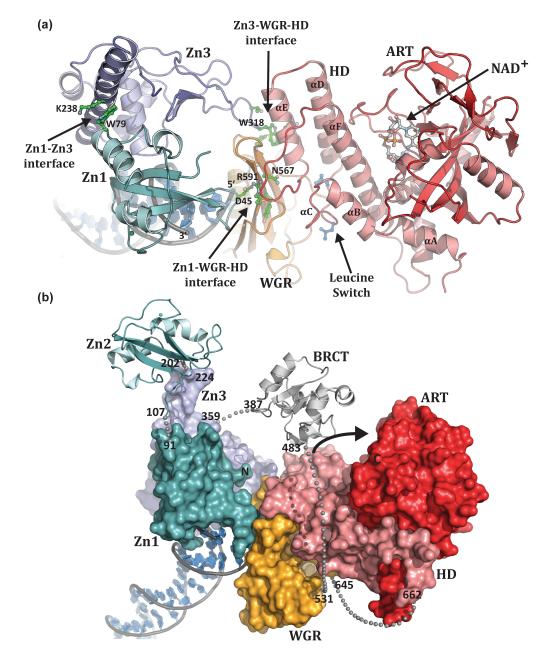
Figure 3. PARP-1 domains collectively assemble on DNA damage.
(a). Human PARP-1 domains Zn1, Zn3, and WGR–CAT were crystallized in complex with a DNA double-strand break (PDB code 4dqy [41]). The PARP-1/DNA complex illustrates how DNA damage detection is coupled to structural transitions in the catalytic domain that elevate poly(ADP-ribosyl)ation activity [41]. Three essential interdomain contact regions form upon PARP-1 interaction with DNA: Zn1–WGR–HD, Zn3–WGR–HD, and Zn1–Zn3. Mutations that target has captured interdomain contacts that are relevant to PARP-1 regulation. Interdomain communication with the HD displaces conserved Leu residues from the hydrophobic interior of the HD, leading to destabilization of the CAT that correlates with an elevation in PARP-1 catalytic activity. (b). A model for the approximate positioning of the Zn2 and BRCT domains within the PARP-1/DNA complex. Zn1, Zn3, and WGR-CAT are shown as surfaces, labeled, and colored as in Figure 1a. The Zn2 and BRCT domains are drawn in schematic representation. Their positioning is based on the relative location of the termini of adjacent domains in the structure. The numbering and location of linker residues are shown. The arrow indicates the location of the PARP-1 automodification region near the catalytic active site.