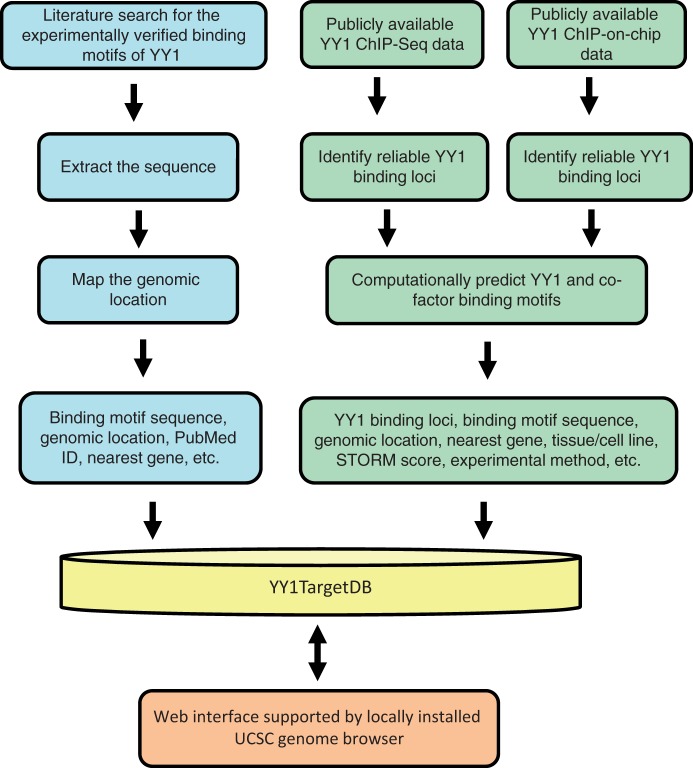
Figure 1.
Schematic overview for YY1TargetDB data acquisition and database implementation. The experimentally verified YY1-binding motifs were collected from literatrues, mapped to the reference genomes and stored in the database. High-throughput (ChIP-seq or ChIP-on-chip) experimental datasets were downloaded from NCBI GEO server and processed by our analysis and annotation pipeline. The identifed YY1-binding loci, computationally predicted YY1 and its cofactors binding motifs were deposited into YY1TargetDB. The database in integrated with locally installed UCSC genome browser to visualize YY1-binding loci and assoicated annotation information.