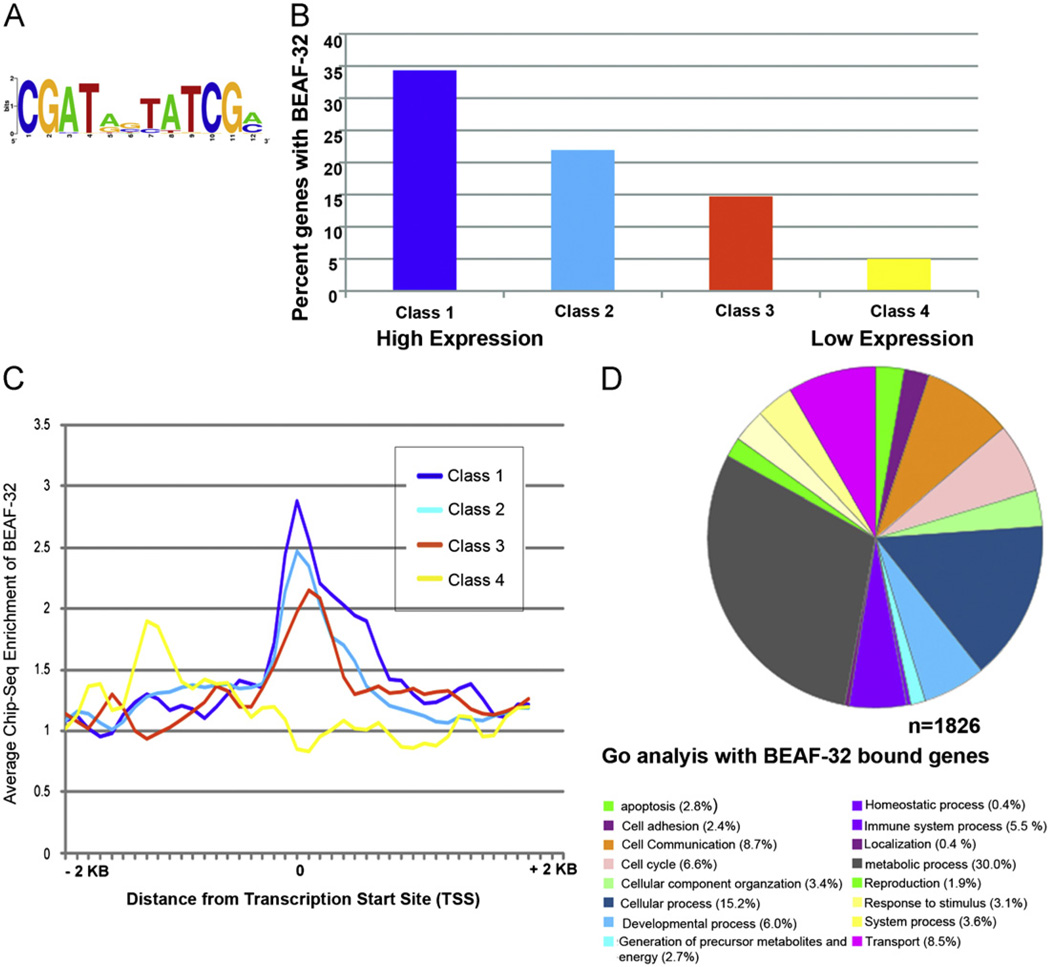
Fig. 3.
Genome wide distribution of BEAF-32 in wing imaginal tissue. (A) BEAF-32 recognition motif predicted using data from ChIP-seq experiments. (B) Histograms indicating the percent distribution of BEAF-32 bound genes and their gene expression levels in wing imaginal tissue. The percentages refer to the fraction of BEAF-bound genes within each class. (C) Distribution of BEAF-32 around transcription start sites in genes with different expression levels. (D) Gene ontology analysis of BEAF-32 bound genes enriched in different cellular processes.