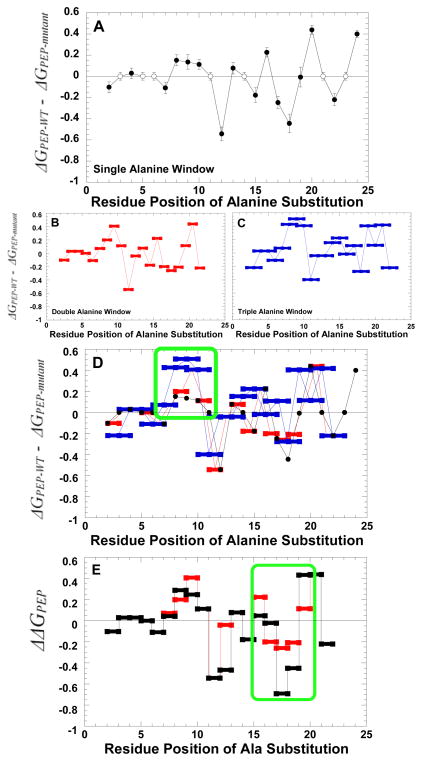
Figure 5.
The impact of an N-terminus alanine scan on the ΔG associated with Ka-PEP. A) The impact of single residues substituted with alanine. Filled circles represent a comparison between measurements for mutant protein with that from wild type protein. Residues that are alanine or glycine in the wild type structure were not probed. Therefore, the wild type data used to represent Ka-PEP at these positions are represented by open circles to clearly distinguish data for mutant proteins. B) The impact of substituting two consecutive positions, both with alanine. C) The impact of substituting three consecutive positions with alanine. D) The overlay of the three data sets included in panels A, B, and C. In all panels, data are compared to that of the wild type protein. A positive deviation from zero on the y-axis indicates reduced PEP affinity; a negative value is improved PEP affinity. Panel D includes both the sum of effects caused by individual mutations (sum of Δ ΔG values for the two single alanine mutations; black) and the difference between the wild type value and that observed for the protein with both alanine substitutions of interest inserted simultaneously (Δ ΔG values for double alanine window mutations: red). As a means of simplifying panels, error bars are only included in panel A. However, error estimates are included for all data in the supplemental material. Green boxes are included in panels D and E to emphasize areas of interest.