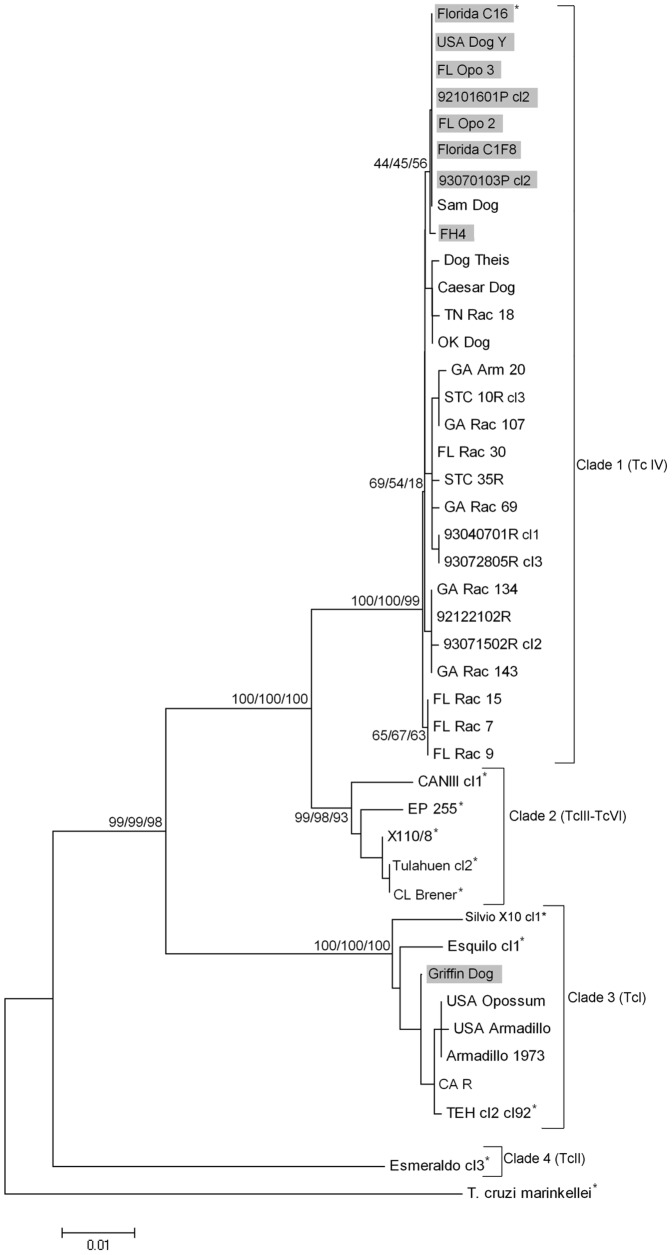
Figure 3. Evolutionary relationships among cytochrome oxidase subunit II- NADH dehydrogenase subunit I region (COII-ND1) from 43 Trypanosoma cruzi isolates.
Three phylogenetic trees were created by neighbor-joining (NJ), minimum evolution (ME), and maximum parsimony (MP) methods from the alignment of each gene target and a consensus tree was interpreted. Numbers at the branches are bootstrap values >50% (500 replicates) for the same nodes of the NJ, ME, MP trees. Evolutionary distances were computed using the Kimura 2-parameter method [29]. The nine isolates with positions incongruent to the nuclear phylogenies (Fig. 2) are highlighted. * = reference strains. Sequences clustered in 4 distinct clades. Clade 1 contains exclusively US origin TcIV strains of T. cruzi. Reference TcIII-TcIV strains of T. cruzi clustered in Clade 2. TcI T. cruzi from the US and S. American reference strains clustered in Clade 3, while a separation of a TcII reference strain results in the fourth clade.