Figure 1. Interaction of Pit2 and the maize protease CP2.
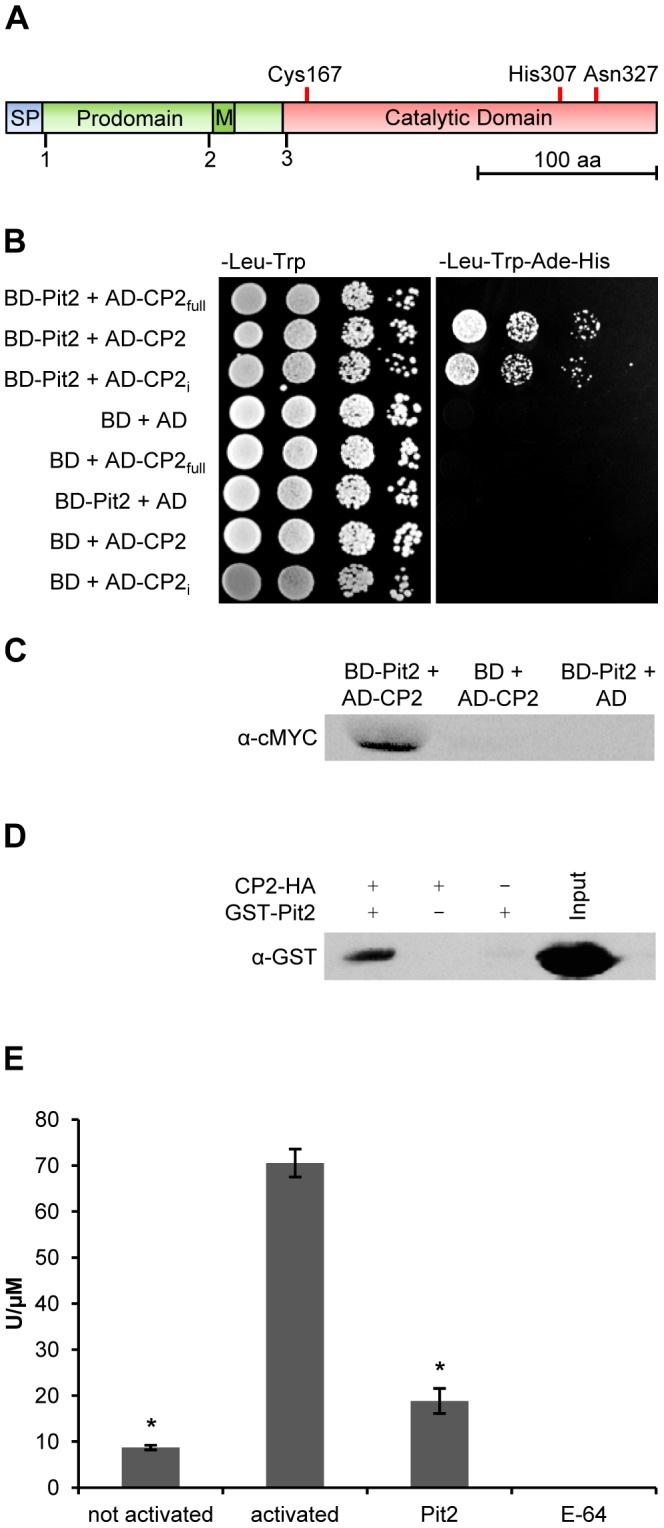
(A) Domain structure of the maize cysteine protease CP2, which was identified as Pit2 interaction partner in a yeast-two-hybrid screen. SP: signal peptide, M: minichain, typical for aleurain-like proteases [29]. Cys167, His307, Asn327 form the catalytic triad. Numbers (1–3) indicate sequence start of CP2 constructs (1) used for heterologous CP2 expression in E. coli, (2) used for Y2H experiments, (3) the CP2 fragment identified in the initial Y2H library screen. (B) Y2H interaction tests of Pit2 with different versions of CP2. Pit2 does not interact with CP2full (Full-length protease including N-terminal prodomain, corresponding to (1) shown in (A)) but interacts with inactivated CP2i (Catalytic triad residues were replaced by Glycine). BD: Gal4 Binding Domain. AD: Gal4 Activation Domain. (C) Co-immunoprecipitation shows interaction of Pit2 and CP2 fusion-proteins isolated from yeast cells. (D) Co-immunoprecipitation of E. coli expressed GST-Pit2 and HA-tagged CP2 protein that was expressed in N. benthamiana (see Figure 3). (E) Fluorescence based protease assay shows activity of recombinant CP2 that was purified from E. coli and its inhibition by Pit2 as well as the specific small-molecule cysteine protease inhibitor E-64. not activated: purified CP2 shows only very low activity. activated: activity of CP2 after activation by pH-shift and treatment with 10 mM pepsin (see material and methods for further details). Pit2: Addition of 10 µM Pit2 resulted in significant reduction of CP2 activity. 2.5 µM E-64 inhibited CP2. The experiment was carried out in three independent replicates; error bars represent SEM; P values were calculated by an unpaired t test. *P<0.05.
