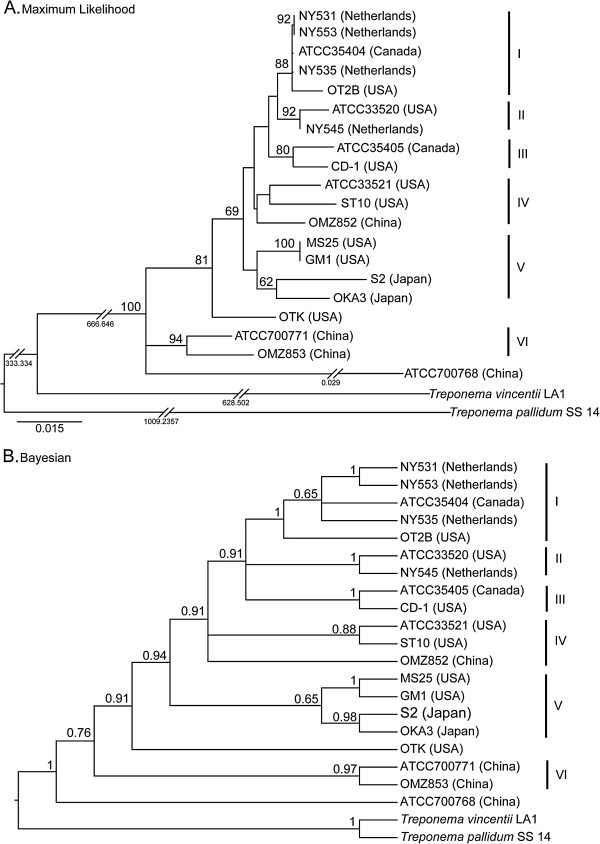
Figure 3.
Phylogenetic trees of Treponema denticola strains based on a concatenated 7-gene dataset (flaA, recA, pyrH, ppnK, dnaN, era and radC), using Maximum Likelihood and Bayesian methods.A: Maximum likelihood (ML) tree generated under the GTR + I + G substitution model, with bootstrap values shown above branches. The scale bar represents 0.015 nucleotide changes per site. Numbers beneath the breakpoints in the branches indicate the respective nucleotide changes per site that have been removed. B: Ultrametric Bayesian (BA) 50% majority-rule consensus tree of 9,000 trees following the removal of 1,000 trees as burn-in. Numbers above branches are posterior probabilities. The respective clades formed in each tree are indicated with a Roman numeral (I-VI). Corresponding gene homologoues from Treponema vincentii LA-1 (ATCC 33580) and Treponema pallidum subsp. pallidum SS14 were included in the phylogenetic analysis as outgroups.